Megabase-scale human genome rearrangement with programmable bridge recombinases
Megabase-scale human genome rearrangement with programmable bridge recombinases
Perry, N. T.; Bartie, L. J.; Katrekar, D.; Gonzalez, G. A.; Durrant, M. G.; Pai, J. J.; Fanton, A.; Hiraizumi, M.; Ricci-Tam, C.; Nishimasu, H.; Konermann, S.; Hsu, P. D.
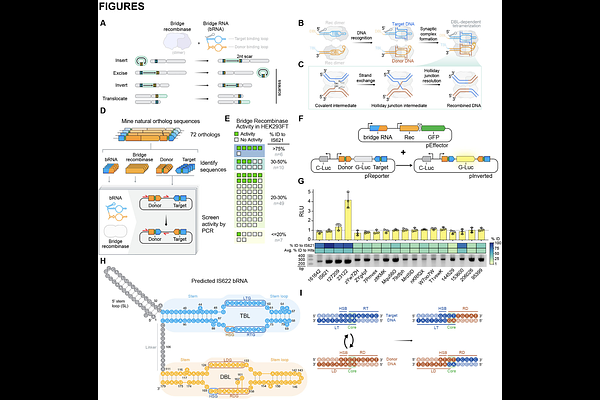
AbstractBridge recombinases are a class of naturally occurring RNA-guided DNA recombinases. We previously demonstrated they can programmably insert, excise, and invert DNA in vitro and in bacteria. Here, we report the discovery and engineering of IS622, a simple and two-component system capable of universal DNA rearrangements of the human genome. We define strategies for the optimal application of bridge systems, leveraging mechanistic insights to improve their targeting specificity. Through rational engineering of the IS622 bridge RNA and deep mutational scanning of its recombinase, we achieve up to 20% insertion efficiency into the human genome and genome-wide specificity as high as 82%. We further demonstrate intra-chromosomal inversion and excision, mobilizing up to 0.93 megabases of DNA. Finally, we provide proof-of-concept for excision of a gene regulatory region or expanded repeats for the treatment of genetic diseases.