Network-based Near-Scalp Personalized Brain Stimulation Targets
Network-based Near-Scalp Personalized Brain Stimulation Targets
Kong, R.; Xue, A.; Ooi, L. Q. R.; Asplund, C.; Tan, X. W.; Goh, S. E.; Lee, J. J.; Koh, J. Z. J.; Tan, R. S. Y.; Singh, H. K. G.; Tan, T. W. K.; Webler, R. D.; Fox, M. D.; Siddiqi, S. H.; Tor, P.-C.; Yeo, B. T. T.
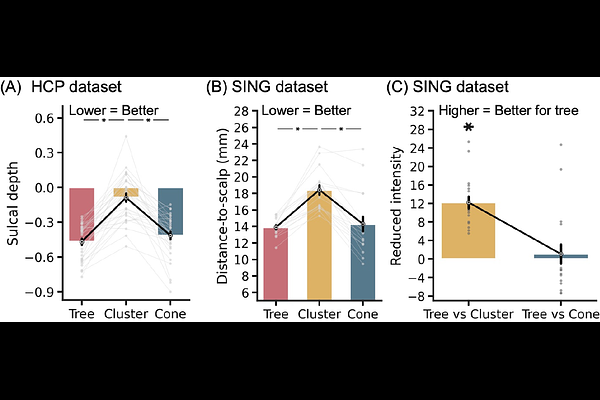
AbstractBackground: Functional connectivity (FC) is used to identify personalized targets for transcranial magnetic stimulation (TMS). However, existing methods often overlook individual differences in whole-cortex network organization and were not explicitly designed to identify TMS targets near the scalp. Objective: We develop a novel framework to simultaneously optimize FC and scalp proximity for personalized target localization. Methods: We use the multi-session hierarchical Bayesian model (MS-HBM) to estimate high-quality individual-specific networks. A tree-based algorithm is then used to select the optimal target location. By essentially having no parameter to tune, our framework might improve generalizability across populations. We compare our approach to existing FC-based algorithms for TMS target selection in terms of scalp proximity, test-retest reliability, and FC to brain regions implicated in depression. Results: In two test-retest datasets of healthy individuals from the United States and Singapore, tree-based MS-HBM reliably identifies personalized TMS targets for depression in close proximity to the scalp. These targets are equivalent or superior to targets identified using prior algorithms in terms of reliability, scalp proximity, and FC to the subgenual anterior cingulate cortex. To demonstrate versatility, we apply the same algorithm, without having to tune any parameter, to identify personalized TMS targets for anxiety. Conclusion: The tree-based MS-HBM algorithm provides a robust, generalizable framework to estimate near-scalp personalized targets across different populations.