Hidden structure in polygenic scores and the challenge of disentangling ancestry interactions in admixed populations
Hidden structure in polygenic scores and the challenge of disentangling ancestry interactions in admixed populations
Aw, A. J.; Mandla, R.; Shi, Z.; Penn Medicine Biobank, ; Pasaniuc, B.; Mathieson, I.
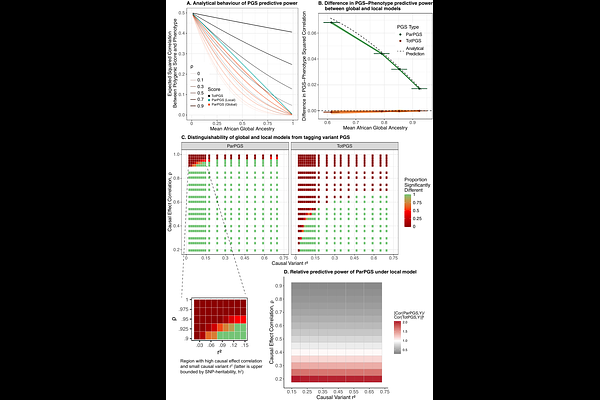
AbstractThe extent to which genetic effects vary across ancestries remains a central question in human genetics, with direct implications for improving polygenic prediction. Recent studies have found that average causal effects are similar across ancestries, but empirical evidence and theoretical considerations suggest that interactions may still contribute to poor portability of polygenic scores. Here, we leverage the ancestry mosaicism of admixed individuals to develop models of genetic interactions with local and global ancestry. We show that these models capture epistasis but are also consistent with highly similar average causal effects, clarifying that the latter observation does not exclude the possibility of gene-ancestry interaction. Next, we investigate how models of local and global interactions differ in polygenic prediction. Focusing on continuous traits, we show that if causal variants are known and causal effects differ across ancestries, then the global and local models can be differentiated using partial polygenic scores --- scores computed on ancestry-specific genomic segments --- while standard polygenic scores remain indistinguishable under both models. However, if causal variants are unknown, differences in linkage disequilibrium (LD) patterns confound this analysis, and the models are not practically distinguishable without knowledge of the LD between causal and tagging variants. Our findings motivate future developments of model-sensitive strategies for individual-level genetic risk prediction.