Bridging Gaps in Soil Ecology: Metagenomic Insights into Microbial Diversity and Functionality Across Brazil's Biomes.
Bridging Gaps in Soil Ecology: Metagenomic Insights into Microbial Diversity and Functionality Across Brazil's Biomes.
Tacca, L. M. A.; Lima, R. N.; Pascoal, P. V.; de Oliveira, M. A.; de Azevedo, C. A. X.; Bambil, D.; Mizotte, P.; Rosinha, G. M. S.; Bittencourt, D. M. d. C. B. M. d. C.; Signor, D.; de Moura, M. S. B.; Trindade, J. P. P.; Volk, L. B. d. S.; Fernandes, F. A.; Lopes, R.; Simoes-Araujo, J. L.; Freire, M.; Rech, E.
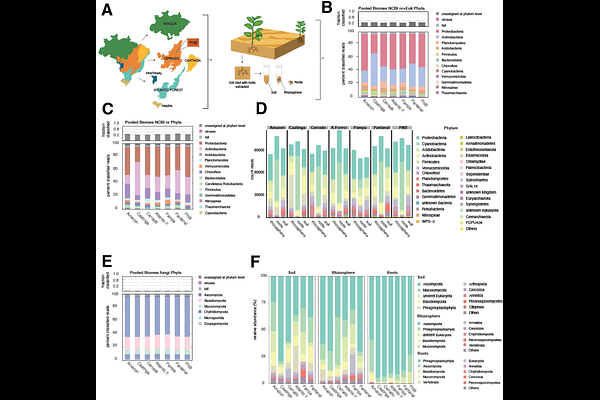
AbstractMicroorganisms participate in complex interactions involving different kingdoms, so rhizosphere biodiversity mapping is essential for understanding how microbes interact with each other in the soil and with roots. Although soil microbial communities are remarkably diverse and technological advances have provided a high capacity to acquire reliable sequence data, unique microbial taxa in soil, root and rhizosphere samples remain poorly described. For the first time, we organized a consortium to collect soil samples covering all Brazilian biomes, providing a comprehensive and unprecedented view of soil microbial diversity. This understanding is critical, especially within the context of climate change, which affects plant physiology, root exudation and, consequently, the composition and functionality of soil microbial communities. The interactions between soil, roots and rhizosphere are influenced by evolutionary and adaptive forces and shape the production of microbial natural products, which exhibit great therapeutic potential and Mapping and studying rhizosphere microbial biodiversity not only increases our knowledge of soil ecology but also offers valuable insights for developing sustainable practices. We employed both 16S/18S/ITS amplicon and metagenomic short-read shotgun sequencing methods to examine and catalogue the large-scale genomes of culture-independent rhizosphere microbes and their interactions with roots in six terrestrial Brazilian biomes, namely, the Amazon, Atlantic Forest, Cerrado, Caatinga, Pampa and Pantanal. Our results revealed the ubiquity of Proteobacteria, which reflects their adaptability to contrasting environments. Biomes with greater moisture availability, such as the Amazon and Pantanal, exhibited greater diversity and abundance of fast-growing bacteria, such as Proteobacteria, and nutrient cyclers, such as Thaumarchaeota. Arid and semiarid biomes, such as the Caatinga, were dominated by microorganisms tolerant to drought and nutrient-limited environments, such as Actinobacteria. Acidobacteria, which thrive in acidic, nutrient-poor soils, were very abundant in forest biomes. The Planctomycetes phylum also occurred more frequently in areas with a relatively high soil organic matter content, such as the Cerrado. Bacteroidetes was significantly more abundant in Pampa than in the other biomes. The results provide comprehensive insights into soil, root and rhizosphere biodiversity and not only enhance the knowledge of the fundamental biological processes sustaining plant life but also constitute a reliable sequencing databank to address present-day agricultural and environmental challenges.