Computational modelling of toroidal membranes at division and fission sites
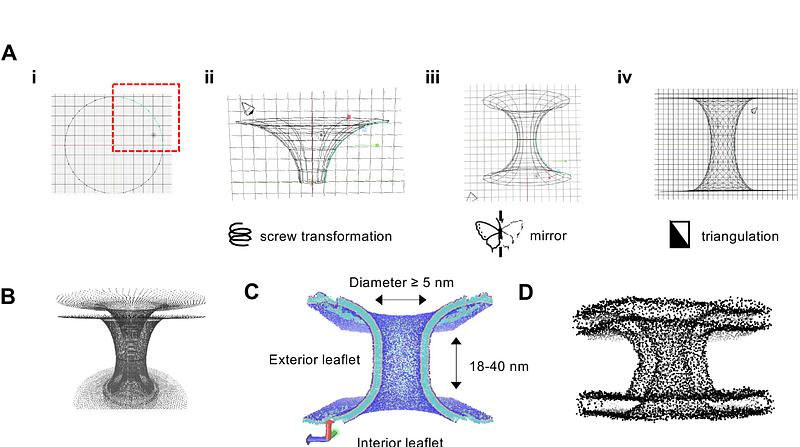
Computational modelling of toroidal membranes at division and fission sites
Graham, C. L.; Parrag, M.; Stansfeld, P. J.; Rodrigues, C. D. A.
AbstractThe formation of specialised membrane architectures is fundamental to biological processes. Computational modelling of membrane structures offers a means to unravel complex molecular mechanisms that remain inaccessible through in vitro or in vivo approaches. Among these architectures, toroidal membranes, otherwise known as fusion pores or fission pores, occur during the final stages of cell division, exocytosis, endocytosis, and membrane fission; including during the phagocytic-like engulfment of bacterial endospores. Here we have designed MemTorMD, a computational simulation pipeline based on the TS2CG methodology, to enable coarse-grained modelling of free toroidal membranes with biologically-relevant dimensions. This approach allowed us to determine the lipid properties required for stable toroidal membrane formation, providing a biologically accurate simulation surface. Furthermore, we demonstrate the applicability of this approach to investigate the function of proteins that localise at toroidal membranes, and drive molecular events such as membrane fission by a prokaryotic fission protein. Collectively, our work provides insight into membrane and protein behaviour at toroidal membranes and provides a platform for the development of more complex simulations involving additional molecular components at membrane toroids present in a range of processes across life.