CladeOScope-GSA: A Tool for Predicting Functional Gene Interactions via Phylogenetic Profiling
CladeOScope-GSA: A Tool for Predicting Functional Gene Interactions via Phylogenetic Profiling
Braun, M.; Bloch, I.; Sherill-Rofe, D.; Canavati, C.; Sharon, E.; Tabach, Y.
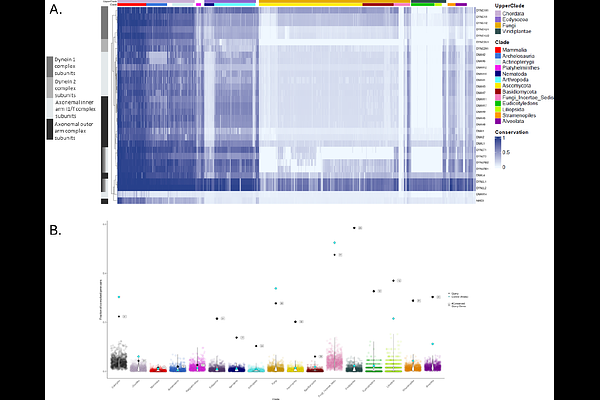
AbstractUnderstanding gene function and protein network dynamics is a key challenge in biology and human health. In the light of the exponential growth in genomic data, phylogenetic profiling methods became more powerful in analysing thousands of genomes to uncover evolutionary associations and predict functional interactions. It is based on the well-supported hypothesis that genes that work together are subject to similar evolutionary pressures and therefore their conservation patterns across phylogeny are correlated. We introduce CladeOScope-GSA, a web-based tool designed for phylogeny-aware analysis of gene sets across extensive comparative genomics datasets. By integrating clade-focused normalized phylogenetic profiling (NPP) with interactive visualization, CladeOScope-GSA enables researchers to identify evolutionary patterns within user-defined gene sets. Building upon the infrastructure of the original CladeOScope visualization platform, the addition of statistical gene set analysis, expanded phylogenetic and genomic data, enhanced visualization, and improved statistical tools represent a significant advancement in the tool\'s capabilities. We demonstrate its utility through case studies spanning 1905 species, showcasing how CladeOScope-GSA empowers researchers to extract biological insights from complex comparative datasets. The web tool is accessible at https://tabachlab.shinyapps.io/CladeOScope