Common misspecification of the generation interval leads to reproduction number underestimation in phylodynamic inference
Common misspecification of the generation interval leads to reproduction number underestimation in phylodynamic inference
Park, Y.; Koelle, K.
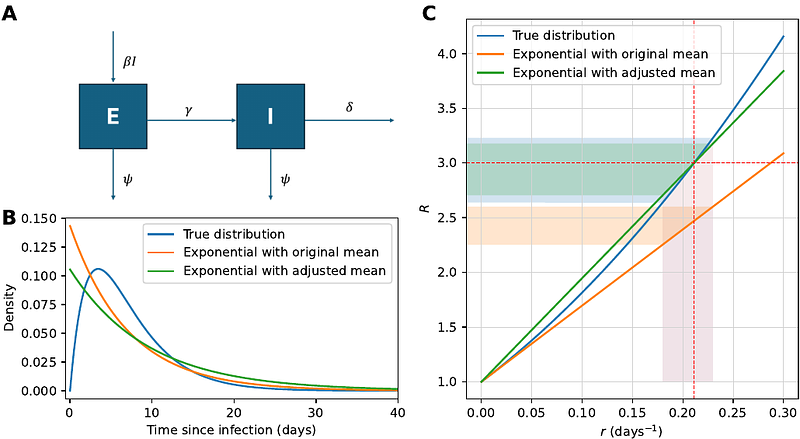
AbstractGeneration intervals are distributions that describe the time between infection and onward transmission. They are a key epidemiological quantity because, together with the reproduction number R, they determine the population-level growth rate of a pathogen and its doubling time. Conversely, when fitting epidemiological models to data, assumed generation intervals impact R inference. This is well-known from studies that have used case data for R inference, with many studies emphasizing the importance of choosing an accurate distribution for the generation interval. In phylodynamic inference of R, the generation interval distribution is often not explicitly mentioned, and the impact of generation interval misspecification has been studied less. Here, we explore the impact of a commonly assumed (but rarely empirically accurate) exponential generation interval distribution on the estimation of R in phylodynamic inference. Using simulations and inference on these simulated datasets, we find that during the early exponential growth of an epidemic, if the generation interval is assumed to be exponentially distributed when it actually has a lower variance, then estimates of R will be biased low. Furthermore, uncertainty in the biased R estimates will be small. Our work highlights the importance of acknowledging implicit generation interval assumptions in phylodynamic inference and points to the need for methodological developments in phylodynamic inference to provide greater flexibility in the specification of accurate generation intervals.