High-resolution genome and genetic map of tetraploid Allium porrum expose pericentromeric recombination
High-resolution genome and genetic map of tetraploid Allium porrum expose pericentromeric recombination
Nieuwenhuis, R.; Voorrips, R.; Esselink, D.; Hesselink, T.; Schijlen, E.; Arens, P.; Cordewener, J.; Scholten, O.; Peters, S.
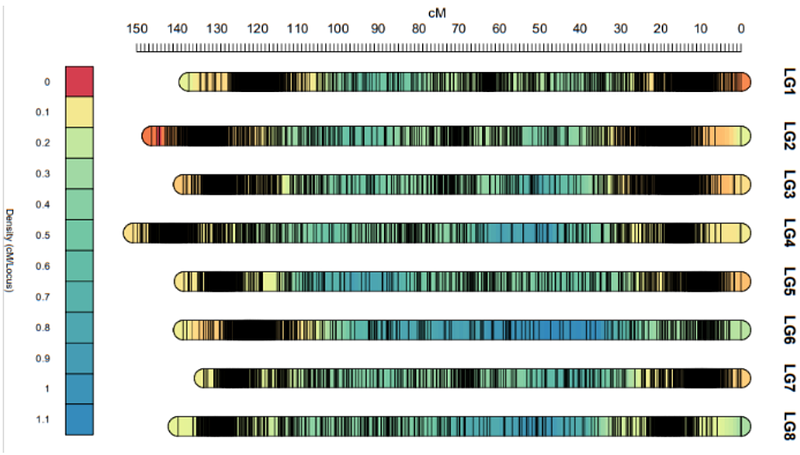
AbstractWe present the first reference genome of highly heterozygous autotetraploid Allium porrum (leek). Combining long-read sequencing with SNP-array screening of two experimental F1 populations, we generated a genetic map with 11,429 SNP markers across 8 linkage groups and a chromosome-scale assembly of Allium porrum (leek) totaling 15.2 Gbp in size. High quality of the reference genome is substantiated by 97.2% BUSCO completeness and a mapping rate of 96% for full-length transcripts. The linkage map exposes the recombination landscape of leek and confirms that crossovers are predominantly proximal located to the centromeres, contrasting with distal recombination landscapes observed in other Allium species. Comparative genomics revealing structural rearrangements between A. porrum and its relatives (A. fistulosum, A. sativum, A. cepa), suggests a closer genomic relationship to A. sativum. Our annotated high-quality reference genome delivers crucial insights into the leek genome structure, recombination landscape, and evolutionary relationships within the Allium genus, offering significant implications for breeding programs, facilitating marker-assisted selection and genetic improvement in leek.