Dissecting serum polyclonal antibody escape to SARS-CoV-2 variants by deep mutational learning
Dissecting serum polyclonal antibody escape to SARS-CoV-2 variants by deep mutational learning
Shlesinger, D.; Sadilek, V.; Minot, M.; Stamkopoulos, E.; Bikias, T.; Kuhn, R.; Agrafiotis, A.; Taft, J. M.; Yermanos, A.; Reddy, S. T.
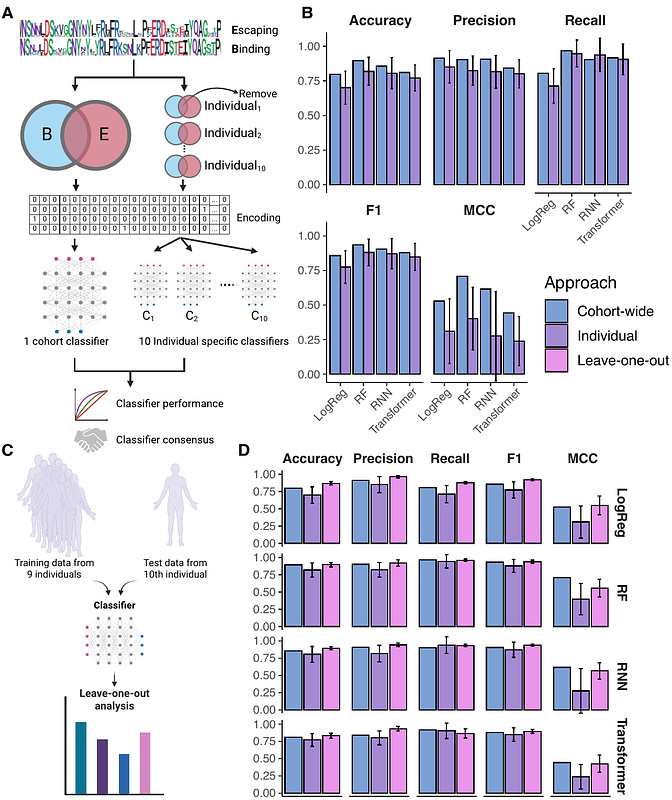
AbstractThe rapid emergence of SARS-CoV-2 variants harboring multiple receptor-binding domain (RBD) mutations continues to challenge the efficacy of vaccines and antibody therapeutics. While deep mutational scanning (DMS) has been instrumental in mapping single-mutation effects on antibody binding and immune escape, it remains limited in its ability to assess combinatorial mutational landscapes. Here, we extend deep mutational learning (DML), a method integrating combinatorial mutagenesis, yeast surface display, deep sequencing, and machine learning, to analyze serum polyclonal antibody escape. Human sera from COVID-19-vaccinated individuals were screened against diverse RBD variant libraries, validating 300 serum-variant interactions across 10 individuals by comparing predicted and observed binding to 30 RBD variants, confirming accurate mapping of binding and escape profiles. Model performance remained consistent across machine learning architectures, suggesting that serum binding and escape are governed by distinct, localized RBD sequence features. Notably, escape profiles were highly individualized, whereas binding signatures were more conserved, reflecting convergent epitope targeting. This approach highlights the potential of DML to generalize beyond observed RBD variants, assess cohort-specific immune breadth, and inform vaccine and therapeutic design in the face of viral evolution.