Phenotypic sex determines recombination rate and distribution in sex-reversed Rainbow Trout Oncorhynchus mykiss
Phenotypic sex determines recombination rate and distribution in sex-reversed Rainbow Trout Oncorhynchus mykiss
Brekke, C.; Knutsen, T. M.
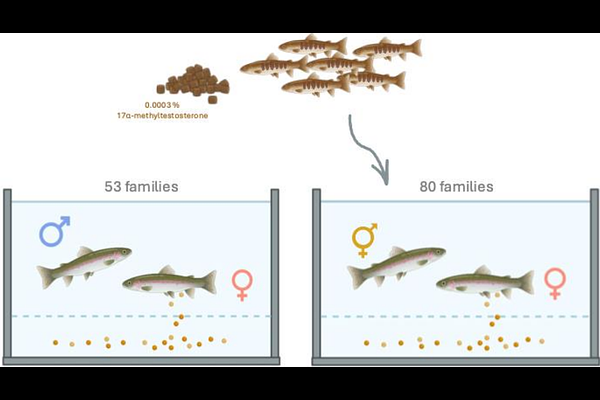
AbstractDuring meiotic cell division, homologous chromosomes align and exchange large segments of DNA through crossover recombination. Rates of recombination often show distinct differences between males and females, a phenomenon known as heterochiasmy. Despite decades of research documenting the presence of heterochiasmy across eukaryotes, the specific feature of sex leading to this curious sexual dimorphism remains to be explained. Some species, such as salmonids, also display extensive differences between the sexes in crossover positioning. A critical part of solving this puzzle is to establish whether heterochiasmy is driven by genetic sex or if it is a result of the physiological differences between producing sperm and eggs. In this study, we show that phenotypic sex determines recombination rate and distribution in hormonally sex-reversed rainbow trout. With pedigree and genotype information from 18 452 individuals and 33 913 SNP markers we map crossover events in families where the fathers were hatched as genetic XX females and sex-reversed as young trout fry with a masculinising hormone 17-methyltestosterone and compare the crossover patterns to those in families with normal XY male fathers. We find that recombination patterns in XX males resemble those of normal XY males with crossovers exclusively in sub-telomeric regions. Crossover count per gamete was 25.8{+/-}4.4 in XX females vs 19.5{+/-}3.9 and 19.9{+/-}4.0 in XY males and XX males, respectively. These results support the hypothesis that heterochiasmy arises from physiological differences between oogenesis and spermatogenesis rather than effects related to genetic sex and will aid in guiding the research on heterochiasmy going forward.