Deep Phenomics of Pattern Recognition Receptor Agonist Specific Activation of Human Blood
Deep Phenomics of Pattern Recognition Receptor Agonist Specific Activation of Human Blood
Barman, S.; Patel, A.; Kelly, A.; Dowling, D. J.
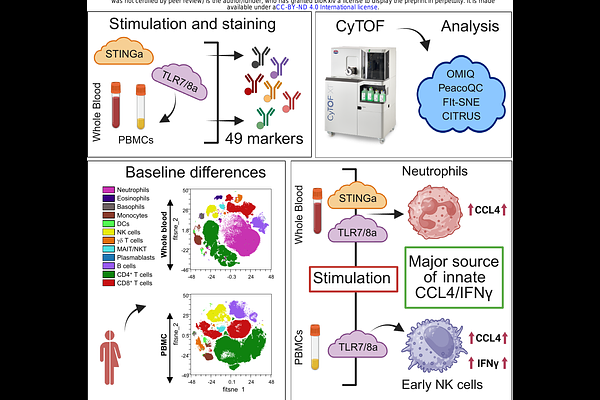
AbstractHuman whole blood (WB) immunophenotyping may represent the in vivo immunological state with better fidelity than artificially isolated peripheral blood mononuclear cells (PBMCs). We used a deep phenomics modeling approach to elucidate the quantitative differences in major immune cell lineages in WB and PBMC compartments in a steady-state in vitro setting. We studied functional innate immune responses induced by pattern recognition receptor agonist adjuvants (PRRa). Using an optimized 49-parameter CyTOF panel and implementing machine learning (ML) algorithms, we mapped PRRa-mediated CD69, CD40, CD80, CD86 and CCR7 activation at the nodal innate immune subsets level. We also portrayed cellular origin of innate functional chemokine CCL4 and intracellular cytokine IFN{gamma} production. We mapped neutrophils as the primary source of TLR7/8 agonist (TLR7/8a) and STING agonist mediated CCL4 responses in WB. Notably, in the PBMC fraction, where neutrophils are limited, natural killer (NK) cells became the major source of innate CCL4 production. TLR7/8a-mediated IFN{gamma} induction by early NK cells was mapped in PBMCs, which was limited in WB. Considering such distinctions, we hypothesized that deep phenomics employing a clinical sample that has not been manipulated, i.e., WB, may be additive in translating in vitro innate fingerprinting into in vivo biology.