High-Resolution Spatial Profiling of Microglia Reveals Proximity Associated Immunometabolic Reprogramming in Alzheimers Disease
High-Resolution Spatial Profiling of Microglia Reveals Proximity Associated Immunometabolic Reprogramming in Alzheimers Disease
Saito, K.; Goulding, D. S.; Nolt, G. L.; Dimas, S. H.; Moore, L. C.; Stevens, I. O.; Anderson, S.; Snipes, A.; Macauley, S. L.; Nelson, P. T.; Johnson, L.; Morganti, J.
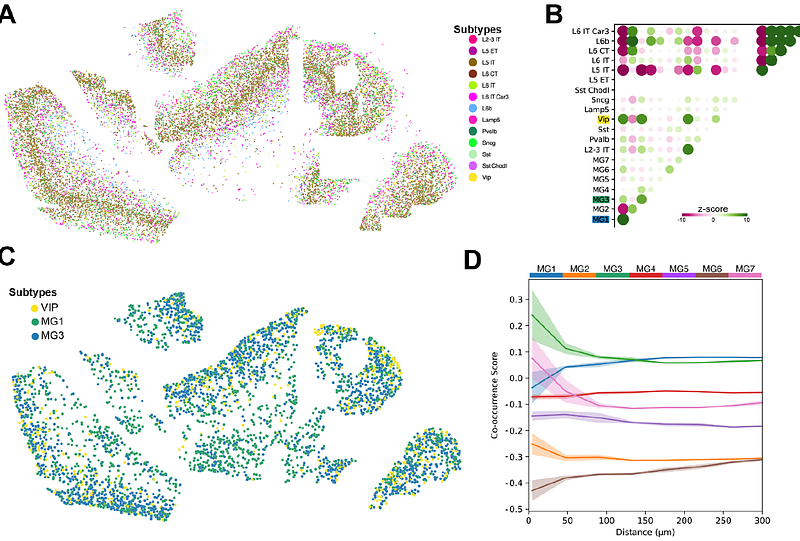
AbstractSingle-cell RNA sequencing has demonstrated that the presence of parenchymal amyloid plaques and intracellular hyperphosphorylated tau pathology is associated with distinctive (and possibly disease-driving) microglial heterogeneity. However, our understanding of how proximity to these Alzheimers disease (AD) pathological hallmarks in situ relates to microglial gene expression remains obscure. Here, we utilized high-resolution spatial transcriptomics (ST) via the Xenium platform with a fully customized gene panel to elucidate disease-associated microglial subtypes in tandem with examining metabolic signatures across AD-relevant mouse models and well-characterized human postmortem tissue. Three mouse models were evaluated: PS19, APP/PS1, and 5xFAD. Analyzing anatomical features across entire hemisections, our approach resolved the distribution of five disease-associated microglial subtypes, while deciphering how proximity to cerebral amyloid plaques influenced transcriptional mediators governing metabolic pathways. We observed robust alterations in glycolytic and cholesterol/lipid processing pathways in plaque-associated microglia, consistent with a specific switch to glycolysis and lipid-fueled metabolism in the plaque niche. Extending our analysis to human postmortem dorsolateral prefrontal cortex (dlPFC), we identified conserved disease-reactive microglial states, i.e., similar proximity-dependent metabolic shifts around amyloid plaques. Further, integrating spatial transcriptomics with machine-learning approaches revealed novel anatomic domain-specific cellular gene expression profiling features, highlighting differential vulnerabilities of neuronal populations near specific microglial subtypes. Together, our findings provide one of the first comprehensive and high-resolution atlas of microglial immunometabolic states across species, anatomical regions, and AD pathological burden.