Evidence-based guidelines for improving network detectability in rodent fMRI
Evidence-based guidelines for improving network detectability in rodent fMRI
Urosevic, M.; Desrosiers-Gregoire, G.; Fouquet, J. P.; Devenyi, G. A.; Gallino, D.; Yee, Y.; Chakravarty, M.
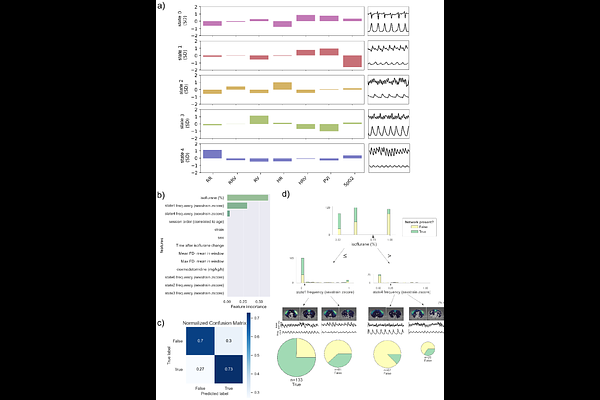
AbstractMouse resting-state functional magnetic resonance imaging (rs-fMRI) is an increasingly popular tool for probing brain activity under experimental manipulations; however, there remains considerable variability in data quality throughout the field. There is a need for an accessible set of acquisition guidelines such that a baseline level of data quality can be attained regardless of domain expertise or specialized equipment (i.e. in anesthetized, free-breathing mice). In particular, there is a gap in the literature regarding the interpretation of physiological parameters as markers of anesthetic depth, and ultimately, data quality. To this end, we developed a set of acquisition guidelines after examining whether continuous physiological variables predict network detectability above and beyond categorical external variables (anesthetic dose, session, time) in C57Bl/7 and C3HeB/FeJ mice anesthetized with isoflurane-dexmedetomidine. Standard physiological metrics (respiration rate and heart rate) did not predict network detectability above and beyond anesthetic dose but instead depended strongly on strain and subject, thus we advise against tuning anesthesia based on respiration or heart rate when the goal is obtaining clear resting-state networks. The most important predictor of improved network detectability was a low isoflurane dose of 0.23%, hence we recommend that researchers prioritize piloting the minimal possible isoflurane dose for their mouse model. In summary, our work examines the contributions from sources of variability that impact rs-fMRI data quality and synthesizes the findings into practical guidelines to help experimenters adapt their acquisition protocols and improve data quality.