ATAC-seq as a versatile tool to portray genomes and epigenomes
ATAC-seq as a versatile tool to portray genomes and epigenomes
Toumi, I.; Lecam, L.; Roux, P.-F.
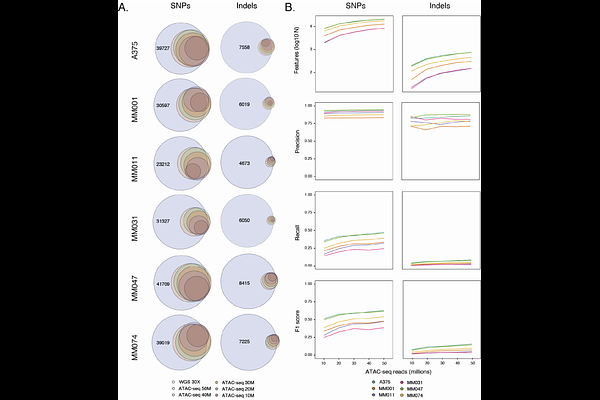
AbstractAssay for transposase-accessible chromatin using sequencing (ATAC-seq) is a cornerstone for epigenomic profiling, yet its potential for genomic characterization remains poorly explored. Here, we systematically benchmarked bulk ATAC-seq against whole-genome sequencing (WGS) to assess its capacity for detecting small variants, copy number variations (CNVs), telomeric content, and mitochondrial mutations. Using paired datasets from six melanoma cell lines, we demonstrated that ATAC-seq achieves reasonable precision in SNP detection, robustly profiles CNVs in the nuclear genome, and effectively quantifies mitochondrial heteroplasmy, with strong concordance to WGS at classical sequencing depths. Notably, we present the first systematic evaluation of telomeric length by ATAC-seq, revealing its untapped potential for studying cellular aging and genome stability. By bridging genomic and epigenomic insights into a single genome-wide approach, bulk ATAC-seq emerges as a cost-effective and versatile tool poised to transform cancer and aging research and advance clinical diagnostics through its integrative capabilities.