Deciphering the Sequence Basis and Application of Transcriptional Initiation Regulation in Plant Genomes Through Deep Learning
Deciphering the Sequence Basis and Application of Transcriptional Initiation Regulation in Plant Genomes Through Deep Learning
Gao, P.; Lian, L.; Feng, W.; Ma, Y.; Lin, J.; Qin, L.; Hao, S.; Zhao, H.; Liu, X.; Yuan, J.; Lin, Z.; Li, X.; Guan, Y.; Wang, X.
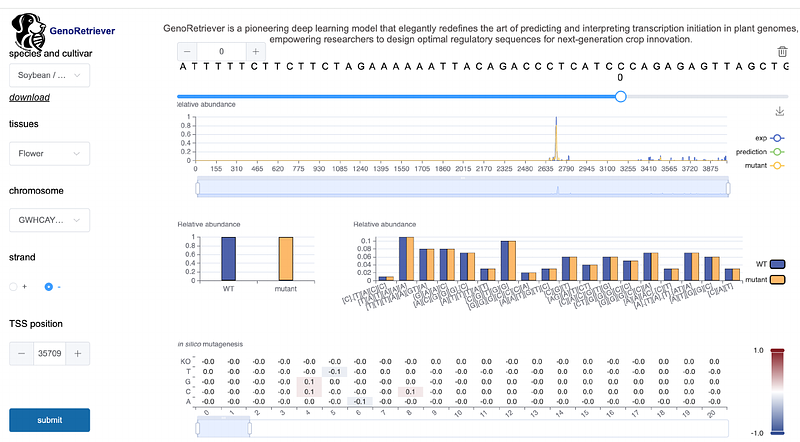
AbstractTranscription initiation is a critical regulatory step in plant gene expression, yet its sequence determinants remain largely elusive. Here we introduce GenoRetriever, an interpretable deep learning model that deciphers the sequence basis of transcriptional initiation regulation across plant genomes. Trained on STRIPE-seq data from 16 soybean tissues and six other crop species, GenoRetriever identifies 27 core sequence motifs that govern transcription start site (TSS) selection and usage. The model predicts TSS locations and usage levels with high accuracy, as validated by in silico motif insertions, saturation mutagenesis, and CRISPR-Cas9 promoter editing. It further reveals that 31.85% of natural variation between wild and domesticated soybean drives shifts in promoter motif usage during domestication, and uncovers lineage-specific motif effects between monocots and dicots. This interpretable model and its user-friendly web server for promoter analysis and design make GenoRetriever both a methodological innovation and practical tool for plant functional genomics and crop improvement.