DNA cross-over motifs based, programmable supramolecular hydrogels for mechanoregulatory effects of cellular behaviour and cytoskeleton reorganization
DNA cross-over motifs based, programmable supramolecular hydrogels for mechanoregulatory effects of cellular behaviour and cytoskeleton reorganization
Singh, A.; Bhatia, D.
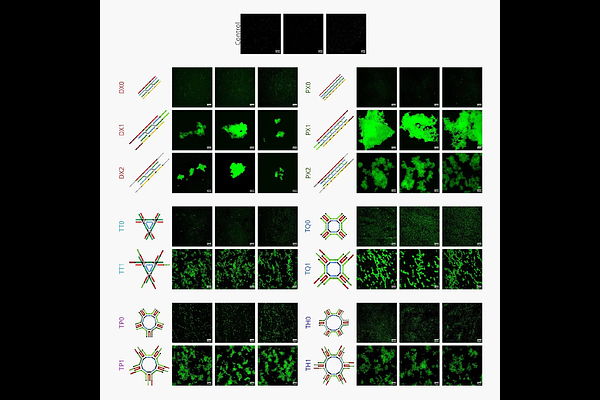
AbstractThe development of mechanically tunable DNA-based supramolecular hydrogels that recapitulate the dynamic mechanoelastic properties of native extracellular matrices (ECMs) is critical for advancing 3D tissue engineering. DNA supramolecular hydrogels with their precise programmable architecture and impeccable control at the nanometre scale assembly make them a highly lucrative biopolymer in the design of ECM-mimicking scaffolds. This study explores the fabrication of stiffness-customizable rigid DNA hydrogels with varying branching architectures, including Double Crossovers (DX), Paranemic Crossovers (PX), and Tensegrity motif structures with three to six arms. Our study reveals that by incorporating different numbers of crossovers, branching arms, and the type of sticky-armed linkers, it is possible to generate a range of DNA hydrogels with varying network geometry and mechanoelastic properties ranging from 50-300 kPa, without any chemical / enzymatic crosslinking aid. These DNA hydrogels mechanoregulate cellular behaviour in retinal pigment epithelial (RPE1) cells by enhancing cellular adhesion, elongation (3 - 8x area increase vs. control), viability (concentration-dependent, 1 - 8x vs. control), organellar homeostasis, including mitochondrial fragmentation and ER stress attenuation. This work establishes a framework for automating DNA hydrogel-based scaffold fabrication processes to meet the preferences of various cells/tissues with bespoke mechanical cues, advancing personalized high-throughput tissue engineering platforms.