Precise MRI-Histology Coregistration of Paraffin-Embedded Tissue with Blockface Imaging
Precise MRI-Histology Coregistration of Paraffin-Embedded Tissue with Blockface Imaging
Wang, Y.; Ho, W.; Huszar, I. N.; DiGiacomo, P.; Taghavi, H. M.; Tao, L.; Choi, M.; Nguyen, N.; Leventis, S.; Camarillo, D. B.; Schlomer, P.; Axer, M.; Shao, W.; Rusu, M.; Cobos, I.; Nirschl, J.; Georgiadis, M.; Zeineh, M.
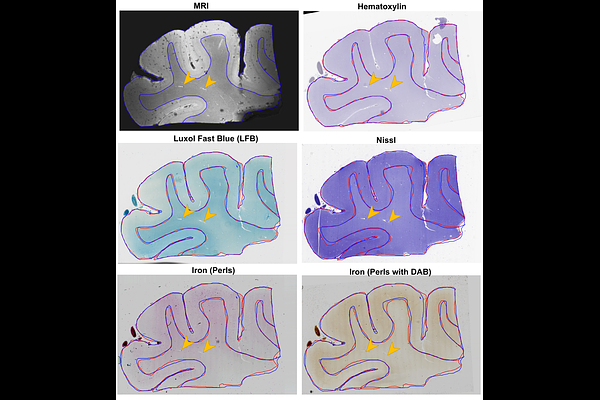
AbstractMagnetic resonance imaging (MRI) provides 3D spatial information on tissue, yet it lacks at the molecular level. In contrast, histology provides cellular and molecular information, but it lacks the 3D spatial context and direct in vivo translation. Coregistering the two is key for the 3D-embedding of histological details, validating pathological MRI findings, and finding quantitative imaging biomarkers of neurodegenerative diseases. However, coregistration is challenging due to non-linear distortions of the tissue from histological processing and sectioning leading to microscopic and macroscopic nonlinear 3D deformations between specimen MRI and stained histology sections. To address this, we developed a novel pipeline, named Brewster\'s Blockface Quantification (BBQ), integrating robust optical approaches with innovative 2D and 3D registration algorithms to achieve precise volumetric alignment of specimen MRI data with histological images. On a variety of brain tissue specimens from distinct anatomical regions and across multiple species, our methodology generated blockface volumes with minimal distortion and artifacts. Using these blockface volumes as an intermediary, we achieve a precise alignment between MRI and histology slides, yielding registration results with an overlapping Dice score of >90% for whole tissue alignment between MRI and blockface volumes, and >95% for 2D MRI-histology registration. This correlative MRI-histology pipeline with robust 2D and 3D coregistration methods promises to enhance our understanding of neurodegenerative diseases and aid the development of MRI-based disease biomarkers.