Spatial heterogeneity of disease infection attributable to neighbor genotypic identity in barley cultivars
Spatial heterogeneity of disease infection attributable to neighbor genotypic identity in barley cultivars
Akram, I.; Rohr, L.; Shimizu, K. K.; Shimizu-Inatsugi, R.; Sato, Y.
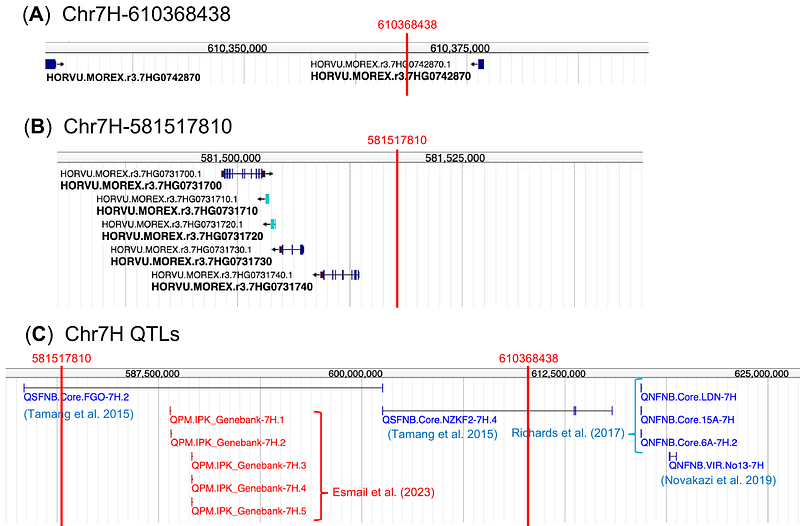
AbstractPest damage exhibits considerable spatial heterogeneity among individual plants in the field. While such spatial heterogeneity has often been treated as a nuisance in crop breeding trials, underlying biotic factors and loci remain poorly understood. To quantify the spatial variation in disease infection and associate it with neighboring genotypes, we applied two methods, Spatial Analysis of Field Trials with Splines (SpATS) and Neighbor Genome-Wide Association Study (Neighbor GWAS), to barley cultivars. Having compiled the CIMMYT Australia ICARDA Germplasm Evaluation (CAIGE) data, we first applied SpATS to three disease phenotypes such as the net form net blotch, spot form net blotch, and scald damage. This SpATS analysis showed extraneous phenotypic variation unexplained by smooth spatial trends, thereby leading us to focus on neighboring genotypes as an extraneous biological factor. We then applied the Neighbor GWAS model and found that neighbor genotypic identity explained 0.10.3 fractional variation in the three disease phenotypes. The Neighbor GWAS method also detected two significant or marginally significant variants on the barley 7H chromosome, which were associated with neighbor genotypic influence on the net form net blotch and scald damage. These variants were estimated to have beneficial effects that reduce disease damage by their allelic mixtures. Our findings suggest that neighbor genotypic identity can account for spatial variation in disease infection, providing a key to reduce pest damage by variety mixtures in field crops.