Solid-State Nanopore Sizing for cfDNA Sample Quality Control in Point-of-Need Sequencing
Solid-State Nanopore Sizing for cfDNA Sample Quality Control in Point-of-Need Sequencing
Khalid, M. A. U.; Ahamed, M. A.; Politza, A. J.; Guan, W.
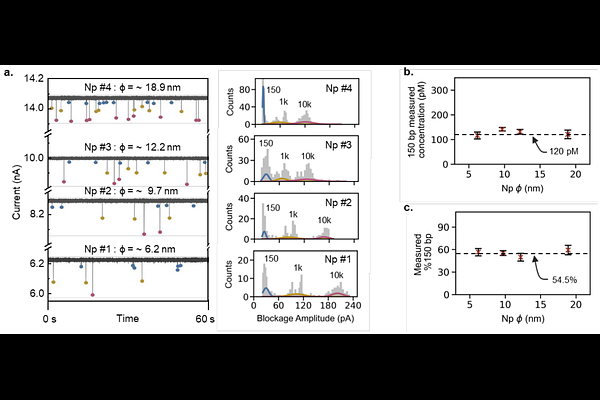
AbstractDNA sequencing is a powerful tool for diagnosing conditions like infectious diseases and cancers. Even though current workflows demand rigorous quality control (QC) of DNA samples, this QC is typically limited to lab settings, despite recent advances in portable nanopore sequencers. For personalized healthcare to truly benefit from the portable sequencer, QC must be performed right where the samples are processed. Here, we present a solid-state nanopore device that provides label-free, controlled quantification and qualification of cell-free DNA (cfDNA). We demonstrated the use of a 1 kbp double-stranded DNA internal marker at a known concentration to measure the concentration of a representative cfDNA target in the presence of genomic DNA. We also found that nanopores with diameters ranging from 6 to 19 nm yield consistent measurements, with a maximum coefficient of variation (CV) of less than 15%. Moreover, analyzing data from multiple nanopores over longer acquisition times can reduce the uncertainty to below 10% CV. Finally, we applied our nanopore QC assay to a plasma cfDNA sample and compared the results with those from a capillary electrophoresis (CE) assay. Both methods produced highly correlated measurements, demonstrating the potential of our nanopore QC assay for effective cfDNA assessment at the point of need.