Single-cell chromatin landscape and DNA methylation patterns reveal shared molecular programs in human tumor and non-tumor tissue CCR8+ Treg cells
Single-cell chromatin landscape and DNA methylation patterns reveal shared molecular programs in human tumor and non-tumor tissue CCR8+ Treg cells
Braband, K. L.; Kaufmann, T.; Beumer, N.; Simon, M.; Helbich, S. S.; Mihoc, D. M.; Voss, M. M.; Nedwed, A. S.; Bauer, K.; Mallm, J.-P.; Ziesmann, T.; Distler, U.; Tenzer, S.; Eckert, C.; Volkmar, M.; Weichenhan, D.; Baehr, M.; Plass, C.; Linke, M.; Diederich, S.; Klein, M.; Bopp, T.; Schild, H.; Henkel, A.; Filippi, E.; Marini, F.; Brors, B.; Gaida, M. M.; Delacher, M.
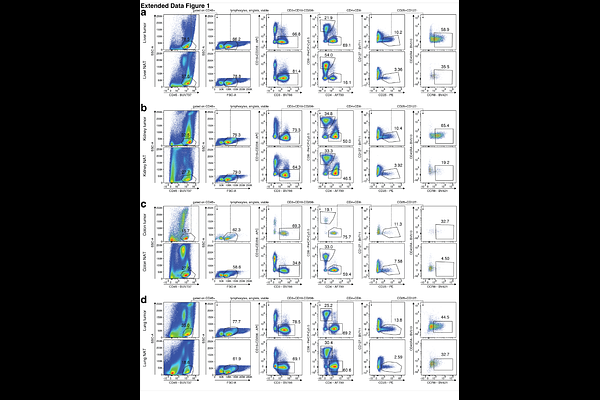
AbstractRegulatory T (Treg) cells, a subset of CD4+ T cells, play a crucial role in immunoregulation. Notably, CCR8-expressing Treg cells in tissues also contribute to organ homeostasis and repair. To determine whether these tissue-regenerative programs are active in the tumor microenvironment, we employed single-cell chromatin accessibility and genome-wide DNA methylation analyses to investigate CCR8+ tissue Treg cells isolated from human tumor and adjacent tumor-free tissues. Our findings indicate that CCR8+ tissue Treg cells from tumor and corresponding tumor-free tissues exhibit a high degree of similarity, suggesting that the tumor microenvironment may harbor highly activated tissue Treg cells. This observation was consistent across various tumor types and origins, including primary tumors and metastases. Using quantitative proteomics, we identified several candidate factors associated with the regenerative and suppressive programs of Treg cells, which may serve as potential reservoir of druggable targets for future therapeutic interventions.