Genomic approaches to build de novo elite breeding gene pools from locally-adapted landraces
Genomic approaches to build de novo elite breeding gene pools from locally-adapted landraces
Fall, S. T.; Kena, A.; Rice, B. R.; Kanfany, G.; Diatta, C.; Kane, N.; Fritz, A. K.; Morris, G. P.
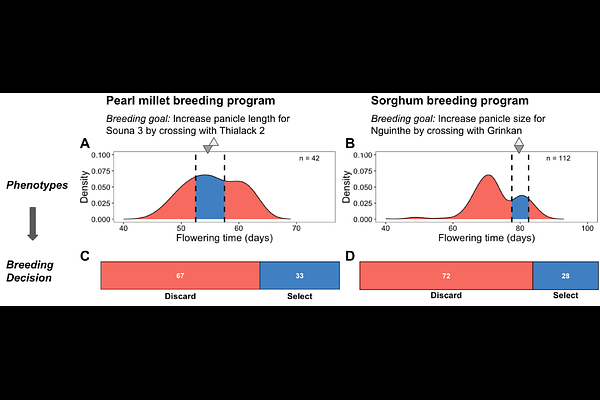
AbstractMany nascent breeding programs aim to achieve genetic gain by crossing locally elite germplasm, but a lack of systematic approaches to develop elite gene pools from locally-adapted varieties hinders their progress. Motivated by the observation of undesirable transgressive segregation in presumed elite crosses in Senegalese cereal breeding programs, we designed approaches for de novo development of elite gene pools from locally-adapted landrace-derived germplasm. We first define two types of \'elite\' germplasm: cis-elite, phenotypically similar and genetically homogeneous for locally-adapted traits (\'acquired traits\'); versus trans-elite, phenotypically similar, but genetically heterogeneous for acquired traits. Next, we defined two genomic approaches for de novo inference of elite gene pools: population-based genotypic inference (PGI) and QTL-based genotypic inference (QGI), and compared to a family-based phenotypic inference (FPI) approach. Using simulations that trace the evolution from locally-adapted landraces to elite breeding lines, we evaluate the effectiveness of these strategies in nascent forward breeding programs. QGI accurately and cost-effectively identifies both cis- and trans-elite pairs, regardless of the underlying trait architecture, while PGI is less sensitive when trait architecture is oligogenic. Over ten cycles of phenotypic recurrent selection, programs based on cis-elite crosses consistently outperformed those based on trans-elite crosses for genetic gain. The findings highlight the value of trait genetic architecture knowledge for elite gene pool development and provide a practical roadmap for elite germplasm development in modernizing breeding programs.