Heritable Large Deletions Using Type I-E CRISPR-Cas3 in Rice
Heritable Large Deletions Using Type I-E CRISPR-Cas3 in Rice
Saika, H.; Hara, N.; Yasumoto, S.; Muranaka, T.; Yoshimi, K.; Mashimo, T.; Toki, S.
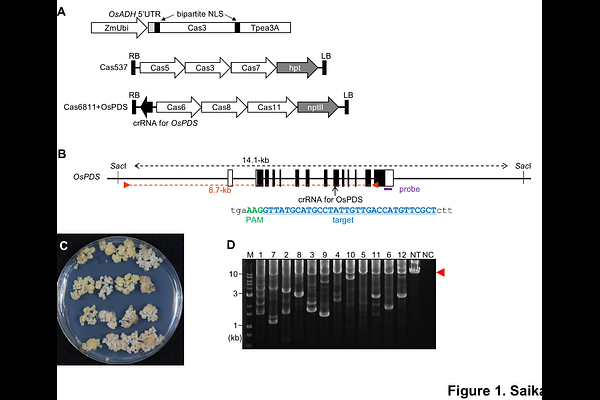
AbstractType I-E CRISPR-Cas3 derived from Escherichia coli (Eco CRISPR-Cas3) can introduce large deletions in target sites and is available for mammalian genome editing. The use of Eco CRISPR-Cas3 to plants is challenging because 7 CRISPR-Cas3 components (6 Cas proteins and CRISPR RNA) must be expressed simultaneously in plant cells. To date, application has been limited to maize protoplasts, and no mutant plants have been produced. In this study, we developed a genome editing system in rice using Eco CRISPR-Cas3 via Agrobacterium-mediated transformation. Deletions in the target gene were detected in 39-48% and 55-71% of calli transformed with 2 binary vectors carrying 7 expression cassettes of Eco CRISPR-Cas3 components and a compact all-in-one vector carrying 3 expression cassettes of Cas proteins fused to 2A self-cleavage peptide, respectively. The frequency of alleles lacking a region 7.0 kb upstream of the PAM sequence was estimated as 21-61% by quantifying copy number by droplet digital PCR, suggesting that mutant plants could be obtained with reasonably high frequency. Deletions were determined in plants regenerated from transformed calli and stably inherited to the progenies. Sequencing analysis showed that deletions of 0.1-7.2 kb were obtained, as reported previously in mammals. Interestingly, deletions separated by intervening fragments or with short insertion and inversion were also determined, suggesting the creation of novel alleles. Overall, Eco CRISPR-Cas3 could be a promising genome editing tool for gene knockout, gene deletion, and genome rearrangement in plants.