AI-based identification of cardiac Purkinje fiber cells isolated from whole adult sheep hearts
AI-based identification of cardiac Purkinje fiber cells isolated from whole adult sheep hearts
Chaigne, S.; Hof, T.; Mimoun, K.; El Ghebouli, A.; Charron, S.; Brette, F.; Choveau, F.; Pasdois, P.; Rucker-Martin, C.; Benoist, D.; Guinamard, R.; Haissaguerre, M.; Walton, R.; Bernus, O.
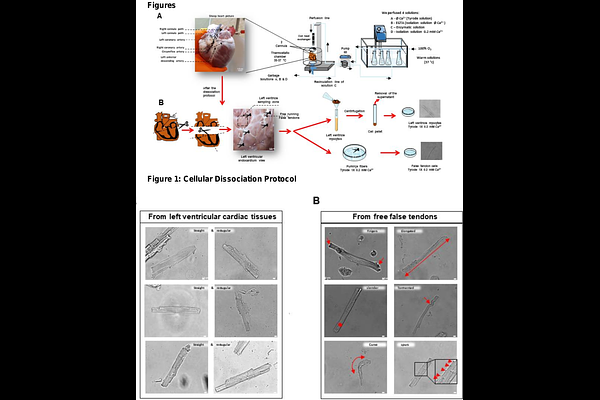
AbstractBACKGROUND: Purkinje Fibers (PFs) are essential to the cardiac conduction system for synchronizing ventricular contractions. However, emerging evidence highlights their implication in the development of ventricular tachyarrhythmias. Nevertheless, isolating and studying the cellular mechanisms of PFs presents a significant challenge due to their intricate arborizing structure, heterogeneous cardiomyocytes (CMs) phenotype, and relatively small proportion within the ventricular mass, all of which hinder detailed functional investigations and comprehensive analysis of the conduction system network. OBJECTIVE: To develop a new methodology for dissociation and classification of cell populations related to the ventricular conduction system from adult sheep. This workflow establishes, in part, a proof-of-concept deep learning-based classification strategy that leverages standard cellular imaging data. METHODS: We developed a multi-tiered workflow to isolate and classify cardiac cell populations from adult sheep hearts. Coronary perfusion and enzymatic digestion were used to dissociate CMs from the left ventricular free wall (LVMs) and Purkinje-rich free-running false tendons (FTs). A three-pronged classification strategy was developed and implemented: (1) expert-guided visual phenotyping based on distinctive morphological traits; (2) rule-based morphometric quantification using automatic image analysis; and (3) deep learning-based classification with a retrained YOLOv9 model trained on augmented brightfield image datasets. This pipeline enabled accurate discrimination between LVM and FT-derived cells. Independent validation was performed using patch-clamp electrophysiology, T-tubule structure imaging with di-8-ANEPPS, and gene expression profiling (RT-qPCR) for Purkinje-specific biomarkers (Tbx5 and Cx40). RESULTS: During the qualitative inspection, FT-dissociated cells had distinct morphological features, including an elongated or slender shape, finger-like projections, curves and tortuous shapes, and a new feature: the presence of spurs along the lateral membrane. Subsequently, a YOLOv9 model achieved an accuracy of 98% in distinguishing LVM and FT cells, based on the initial visual selection made by the operator. In addition, FT-cells exhibit a lower organization and density of T-Tubules compared to LVM. This classification was confirmed by the characterization of the typically longer action potential (AP) durations in FT cells. Finally, higher mRNA expression of the transcription factor Tbx5 and connexin40 (Cx40) was observed in FTs compared to left ventricular tissues. CONCLUSIONS: We present a robust and scalable workflow for isolating and classifying cardiac Purkinje fiber cells from adult sheep, integrating manual phenotyping, rule-based morphometrics, and AI-driven deep learning. This multimodal approach enables high-accuracy identification of PF cells within heterogeneous tissue, confirmed through structural, molecular, and electrophysiological validation. Our findings overcome long-standing barriers in Purkinje fiber research and provide a powerful platform for advancing the study of ventricular