SIMIT-seq: a bead-free, scalable microfluidic platform for single cell mRNA sequencing with ultrahigh cell-indexing rate
SIMIT-seq: a bead-free, scalable microfluidic platform for single cell mRNA sequencing with ultrahigh cell-indexing rate
Duan, M.; Gao, W.; Li, G.; Cai, Y.; Zhao, Z.; Lan, X.; Wang, D.; Xing, X.; Luo, Y.
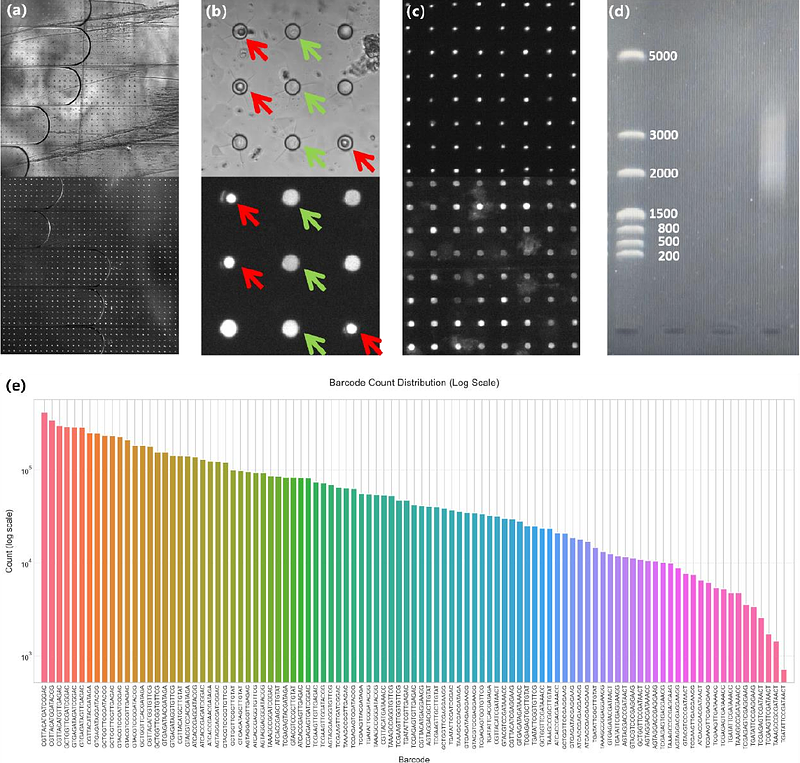
AbstractSIMIT-seq is a bead-free, scalable microfluidic platform designed for high-efficiency single-cell mRNA sequencing. Conventional microfluidic-based single-cell RNA sequencing platforms rely heavily on barcoded beads and intricate co-encapsulation schemes, often constrained by double Poisson limitations and the complexities of bead synthesis. In contrast, SIMIT-seq eliminates the need for beads entirely by employing a deterministic, orthogonal barcoding strategy within a two-dimensional micro-well array. This platform achieves an impressive single-cell indexing rate of 96.6% without the need for complex microfluidic operations. Here, we describe the design and fabrication of the SIMIT-seq platform, outline its workflow for transcript capture and library preparation, and demonstrate its application in profiling K562 cells. Our results validate both the high fidelity of molecular barcode immobilization and the platform capability to support downstream single-cell mRNA sequencing. SIMIT-seq offers a cost-effective and scalable alternative for single-cell transcriptomics, providing a promising foundation for future single-cell omics applications.