High-Sensitivity Top-Down Proteomics Reveals Enhanced Maturation of Micropatterned Induced Pluripotent Stem Cell-Derived Cardiomyocytes
High-Sensitivity Top-Down Proteomics Reveals Enhanced Maturation of Micropatterned Induced Pluripotent Stem Cell-Derived Cardiomyocytes
Wilson, M. C.; Josvai, M.; Walters, J. K.; Lawson, J.; Rossler, K. J.; Gao, Z.; Zhu, Y.; Kamp, T. J.; Crone, W.; Eckhardt, L. L.; Ge, Y.
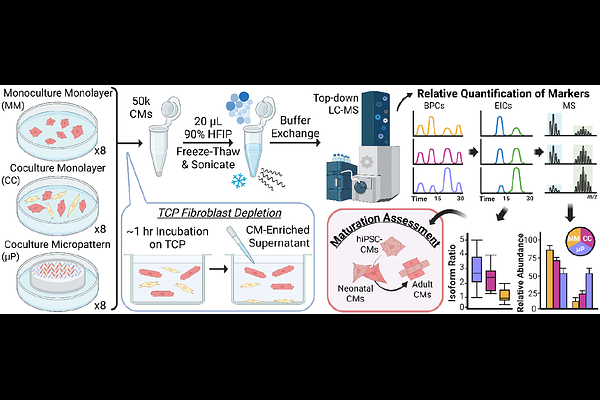
AbstractHuman induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are increasingly used for disease modeling, drug discovery, and precision medicine, yet their utility is often limited by their immature phenotype. One promising maturation strategy involves using micropatterned substrates that mimic native cardiomyocyte organizational growth and stiffness. However, the molecular maturity of this model has not yet been assessed, and there is currently no method to extract proteins from micropatterned hiPSC-CMs for top-down proteomic analysis. Herein, we present a high-sensitivity, surfactant-free protein extraction protocol for the top-down proteomic analysis of hiPSC-CMs. Through this method, we assessed the maturation of micropatterned hiPSC-CMs compared to traditional unstructured monoculture and co-culture monolayers at the proteoform level. We found that micropatterned hiPSC-CMs display molecular signatures of cardiomyocyte maturations including increased expression of ventricular myosin light chain isoforms, reduced expression of the fetal troponin T isoform, and decreased phosphorylation of alpha-tropomyosin. This surfactant-free, high-sensitivity approach enables robust top-down proteomics from limited, heterogeneous cell populations, and identifies the micropattern hiPSC-CM as a more adult-like CM model, broadening the utility of structured culture systems for cardiac disease modeling and translational research.