Novel insights into the genome organization of Rhizobiaceae: identification of linear plasmids
Novel insights into the genome organization of Rhizobiaceae: identification of linear plasmids
Kuzmanovic, N.; Wick, R. R.; Kalita, M.; Pulawska, J.; DiCenzo, G. C.; Hynes, M. F.; Smalla, K.; Babin, D.; Thuenen, T.
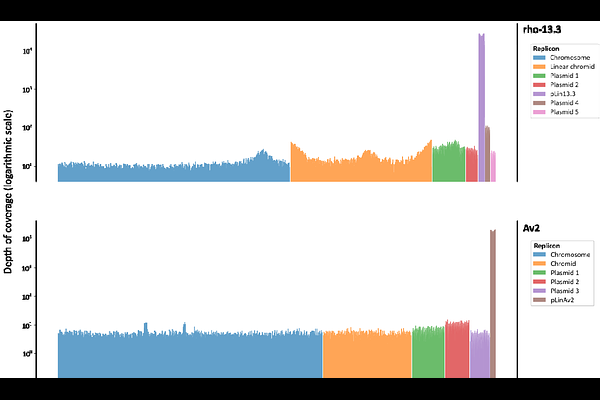
AbstractMembers of the family Rhizobiaceae typically have multipartite genomes, that are split between two or more replicons, including the chromosome and a variable number of extrachromosomal replicons (chromids and plasmids). Nearly all Rhizobiaceae replicons sequenced and described to date have a circular topology, with the exception of the linear chromid found in the genomes of most Agrobacterium spp. In this study, genomes of five nonpathogenic Agrobacterium strains and one plant tumorigenic Allorhizobium strain were fully sequenced. Surprisingly, genome analysis revealed that these six strains each carry an 80-kbp linear plasmid. Linear plasmids were so far not identified in this bacterial family or other bacteria within the class Alphaproteobacteria. The ends of all six plasmids identified in this study have a hairpin structure with covalently closed ends. The plasmid sequences showed a high degree of homology, clearly indicating their common ancestry. Database searches led to the identification of additional linear plasmids in previously published Rhizobiaceae genome assemblies that were not previously recognized to have linear plasmids, suggesting that these replicons may be more widespread. Most likely, linear plasmids may be even more widely distributed than anticipated. Although the biological functions of the linear plasmids identified in this study remain unknown, they are associated with both nonpathogenic and plant tumorigenic Rhizobiaceae strains.