Protocol for the development and use of spike-in control for chromatin immunoprecipitation (ChIP) of chromatin-binding proteins
Protocol for the development and use of spike-in control for chromatin immunoprecipitation (ChIP) of chromatin-binding proteins
Khanduja, J. S.; Motamedi, M.
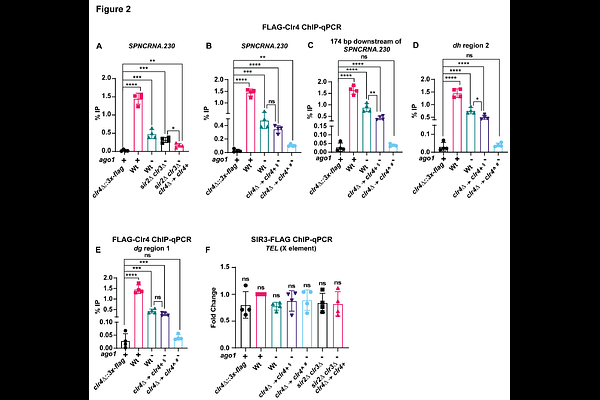
AbstractChromatin immunoprecipitation (ChIP) assays provide quantitative information about the genomic localization of chromatin-binding proteins. However, their sensitivity is limited by several technical variables. To generate high-confidence datasets, in this protocol, we used the Saccharomyces cerevisiae chromatin as an exogenous spike-in control for the ChIP of two S. pombe heterochromatin-associated proteins. This permitted normalization of the ChIP signals based on immunoprecipitation efficiencies across samples. Here, we describe the steps for spike-in control preparation, validation, and its use in data normalization. For complete details on the use and execution of this protocol, please refer to Khanduja et al.1