Streamlined isolation of the n-terminome via phosphonate tagging and coordination-based depletion
Streamlined isolation of the n-terminome via phosphonate tagging and coordination-based depletion
Giansanti, P.; Fojnica, A.; Pichlmair, A.
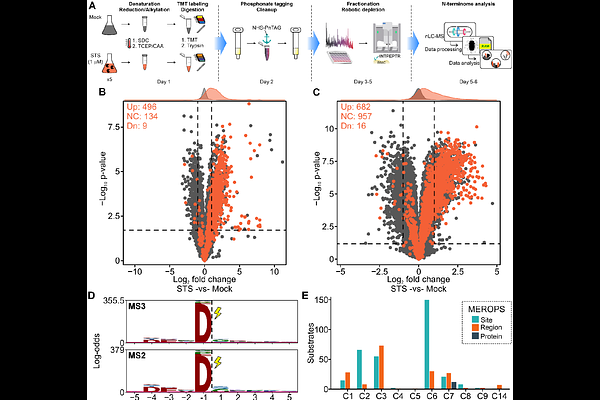
AbstractProtein degradation is critical for regulating cellular functions. Therefore, there is considerable interest in developing effective proteome-wide strategies to identify protease cleavage products to enhance our understanding of proteolytic pathways and their perturbation in diseases. Here, we present a streamlined N-termini proteome analysis leveraging N-Hydroxysuccinimide (NHS) ester chemistry. At the protein level, N-terminal amines (naturally occurring protein N-termini, lysines, or protease-generated N-termini) are blocked directly in the cell lysate, and tryptic digestion is performed straight after, without the need for any buffer exchange. The internal tryptic peptides are tagged with a phosphonate moiety and subsequently depleted via metal-based coordination, keeping exclusively N-blocked terminal peptides for analysis by LC-MS/MS. We demonstrate the applicability of this approach by monitoring proteolytic events induced by intrinsic apoptotic pathway. Our approach identified more than 690 cleaved proteins in response to Staurosporine-induced apoptosis, including many previously unknown substrates and cleavage sites. The presented approach is, therefore, a straightforward and robust method for mass spectrometry-based identification of caspase-generated cleavage products and extendible to a wide range of other proteolytic cleavage events.