TransMetaSegmentation (TMS): a transcriptome-based segmentation method for spatial metabolomic data
TransMetaSegmentation (TMS): a transcriptome-based segmentation method for spatial metabolomic data
Wang, Y.; Bender, K. J.; Zhang, W.; Lin, S.; Neumann, E. K.; Wang, A.
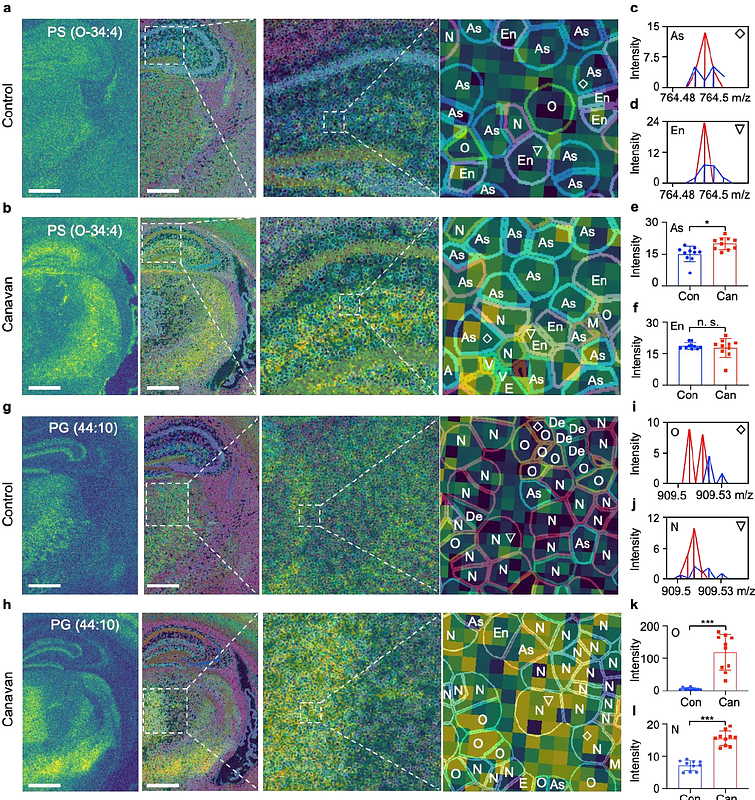
AbstractMatrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) is a powerful analytical tool that enables the visualization and comparison of relative abundances of metabolites across samples, shedding light on biological processes and disease mechanisms. Techniques such as scSpatMet enable the determination of cell boundaries and cell types through staining with 35 cell marker antibodies. Yet, distinguishing subpopulations of cells, such as astrocytes, oligodendrocytes, and neuronal clusters in the brain, remains challenging using antibodies. In this context, we introduce TransMetaSegmentation (TMS), an alternative segmentation and cell typing method that integrates MALDI MSI imagery with single-cell spatial transcriptomic analysis. This approach not only delineates cell boundaries and defines cell types based on a number of marker genes but also effectively allocates metabolites to specific cell types in a high-throughput manner. We anticipate that TMS will improve the granularity of MALDI MSI analyses, advance our understanding of metabolic alterations in diseases, and have an impact on various fields within biomedical sciences.