Advanced eTAM-seq enables high-fidelity, low-input N6-methyladenosine profiling in human cells and embryonic mouse tissues
Advanced eTAM-seq enables high-fidelity, low-input N6-methyladenosine profiling in human cells and embryonic mouse tissues
He, E. M.; Wu, Y.; Ye, C.; Zeng, T.-B.; He, C.; Tang, W.
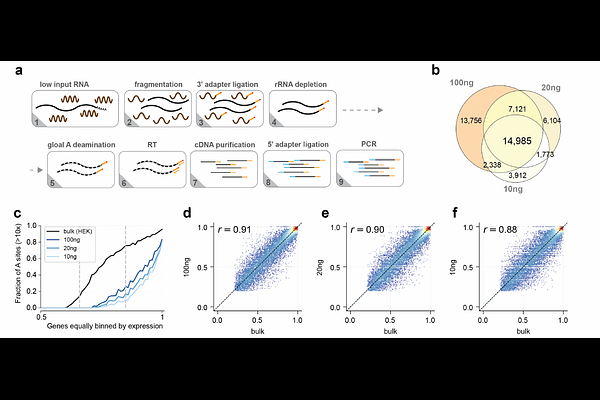
AbstractFunctional dissection of N6-methyladenosine (m6A), the most abundant internal messenger RNA modification in mammals, demands quantitative, scalable detection technology. We previously reported eTAM-seq, which supports transcriptome-wide quantification of m6A by enzyme-assisted adenosine deamination. While effective, TadA8.20--the enzyme used in our first-generation technology--is sensitive not only to m6A but also to RNA structure, making accurate detection dependent on the inclusion of control transcriptomes. Here, we introduce eTAM-seq-v2, in which we replace TadA8.20 with TadA8r, a further evolved adenosine deaminase with superior catalytic efficiency. eTAM-seq-v2 supports control-free m6A calling with high fidelity. Because enzyme treatment preserves RNA integrity, eTAM-seq surveys >51% of A sites in all expressed genes with moderate sequencing depth (60 million uniquely mapped reads) and delivers robust performance with as little as 10 ng of total RNA (~500 cells). With eTAM-seq-v2, we delineate the m6A landscape across six human cell lines and seven embryonic mouse tissues. While uncovering broadly conserved m6A patterns, we reveal that most neighboring m6A sites are independently deposited at the single-molecule level. Moving forward, we envision that eTAM-seq-v2 will enable researchers to survey m6A in diverse biological contexts and uncover new insights into its regulatory roles.