Nucleosome positioning and sensitivity suggest novel functional organization of the purple sea urchin (Strongylocentrotus purpuratus) genome
Nucleosome positioning and sensitivity suggest novel functional organization of the purple sea urchin (Strongylocentrotus purpuratus) genome
Munstermann, M. J.; Benoit, J. M.; Karelitz, S. E.; Arambarri, L. N.; Dennis, J. H.; Okamoto, D. K.
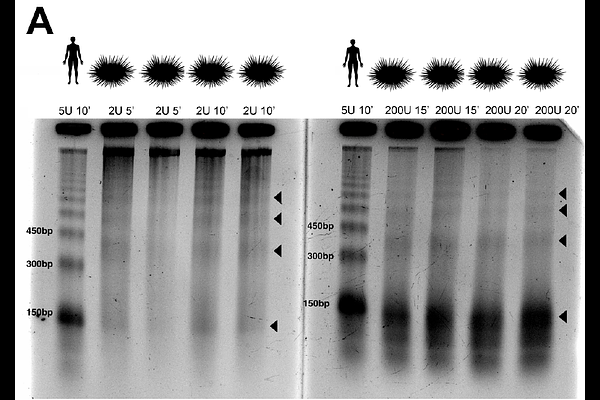
AbstractThe eukaryotic nucleosome is the fundamental subunit of chromatin and plays functional roles in DNA templated processes, including replication and transcription. In eukaryotic promoters, nucleosome organization is highly structured, with nucleosomes occupying canonical positions flanking the transcription start site (TSS), thereby regulating access of the transcriptional machinery to the underlying DNA. We determine whether this canonical distribution is present in the purple sea urchin, Strongylocentrotus purpuratus, a species of ecological importance and a model organism for developmental biology and climate science. We used titrations of micrococcal nuclease to produce high throughput maps of nucleosome distribution and sensitivity to digestion from female urchin gonad tissue. Unlike yeast, flies, zebrafish, maize, mice or humans, urchins have extended nucleosome repeat lengths and lack a nucleosome depleted region over TSSs. Urchin promoters are dominated by strongly positioned and highly occupied +1 and +2 nucleosomes which are most prominent in highly expressed genes. Additionally, urchin promoters exhibit distinct patterns of susceptibility to nuclease digestion, with heightened sensitivity upstream of the TSS and limited resistance to nuclease digestion. Discretely positioned sensitive nucleosomes were enriched in promoters of highly expressed genes, suggesting a relationship between nucleosome sensitivity and transcriptional regulation. Collectively, we present a comprehensive overview of the unique interplay between nucleosome positioning and chromatin sensitivity in sea urchins. Our study not only provides a better understanding of dynamics of gene expression in a key developmental organism, but also reveals potential heterogeneity in a key structural property of chromatin previously thought to be homogeneous in model eukaryotes.