Megabase-scale detachment of genome-nuclear lamina interactions is required for efficient repair of double stranded breaks
Megabase-scale detachment of genome-nuclear lamina interactions is required for efficient repair of double stranded breaks
Vergara, X.; Manjon, A. G.; Novais-Cruz, M.; de Haas, M.; Ouchene, A.; van Schaik, T.; Manzo, S. G.; Cardoso da Silva, R.; Baubec, T.; van Steensel, B.; Medema, R. H.
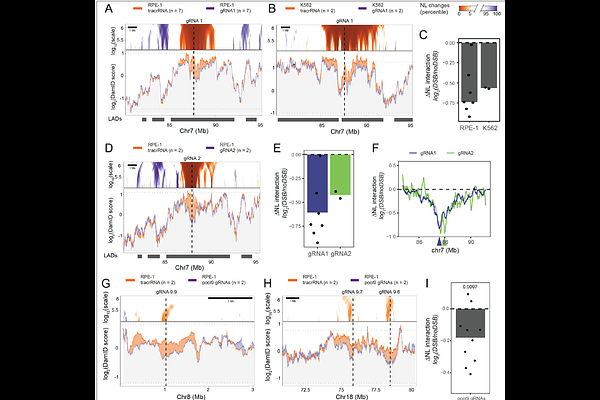
AbstractLamina-associated domains (LADs) are large genomic regions that contact the nuclear lamina (NL). Double-strand breaks (DSBs) in LADs are known to be repaired more slowly and with different pathway preferences compared to other chromatin contexts. However, little is known about the chromatin changes at LADs that occur during DSB repair. Here, we report that a single DSB inside a LAD can cause detachment from the NL over several megabases. This profound spatial rearrangement is transient and reverts within 48 hours. Preventing this detachment slows down repair kinetics and renders repair incomplete, indicating that NL detachment is required for efficient repair of DSBs in LADs. NL detachment is dependent on {gamma}H2AX and ATM, while it is antagonized by DNAPKcs activity. Remarkably, {gamma}H2AX also antagonizes NL interactions at chromosome ends. Taken together, our data indicate that {gamma}H2AX accumulation in LADs induces large scale rewiring of genome-NL interactions, allowing for efficient repair of DSBs.