Effect of Chronic Stress on Whole Blood Transcriptome: A Meta-Analysis of Publicly Available Datasets from Rodent Models
Effect of Chronic Stress on Whole Blood Transcriptome: A Meta-Analysis of Publicly Available Datasets from Rodent Models
Flandreau, E. I.; Nguyen, D. M.; Hagenauer, M. H.; Nguyen, M.; Kim, H.; Duan, T.; Bader, A.; Watson, S.; Akil, H.
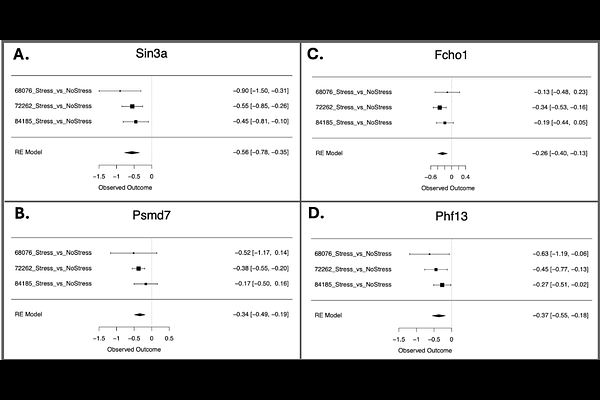
AbstractBackground: Chronic stress increases risk for neuropsychiatric disorders in humans. By modeling stress-induced changes in animals, we may improve diagnosis or treatment of these disorders. Successful translation benefits from studies with sufficient statistical power and outcome measurements that can be directly compared across species. We performed a meta-analysis to examine the impact of chronic stress on the whole blood transcriptome. Methods: Datasets were systematically identified in Gemma, a database of reprocessed public transcriptional profiling studies; datasets GSE68076, GSE72262, and GSE84185 met inclusion/exclusion parameters. Each study exposed eight-week old mice to chronic stress (5-10 days social defeat stress or 6-8 weeks chronic mild stress). The final sample size was n=92 (n=45 Non-Stress/n=47 Stress). Stress-related differential expression in each dataset was quantified using the Limma pipeline followed by empirical Bayes moderation. For the 9,219 genes represented in all three datasets, we ran a meta-analysis of Log(2) Fold Changes using a random effects model and corrected for false discovery rate (FDR). Functional patterns were assessed with fast Gene Set Enrichment Analysis. Cell type specific enrichment for each of the differentially expressed genes was further explored using a public 10x genomics scRNA-Seq dataset from mouse peripheral blood mononuclear cells. Results: Findings included 23 downregulated and 16 upregulated transcripts in stress-exposed mice (FDR<0.05). Results indicated a down-regulation in gene sets related to B cells, immune response, DNA and chromatin regulation, ribosomal activity, translation, and catabolic cellular processes. Upregulated gene sets related to erythrocytes and oxygen binding. Conclusion: Our results provide molecular insight into stress-related immune dysregulation and add weight to the hypothesis that environmental stress escalates cellular aging, supporting the use of blood transcriptome as a bridge between human and rodent models.