Sequence-Dependent Dynamics of U:U Mismatches in RNA Revealed by Molecular Dynamics Simulations
Sequence-Dependent Dynamics of U:U Mismatches in RNA Revealed by Molecular Dynamics Simulations
Krepl, M.; Knappeova, B.; Sponer, J.
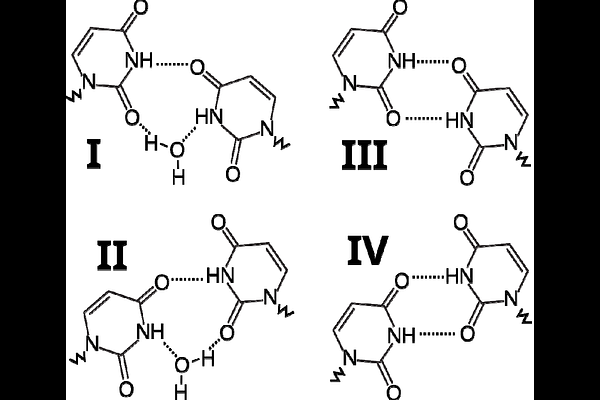
AbstractThe uracil:uracil (U:U) base pair is one of the most common mismatches observed in RNA. It is notable for its ability to adopt multiple conformational states depending on its structural environment, particularly on the identity of the flanking canonical base pairs. Here, we employed extensive molecular dynamics (MD) simulations to systematically investigate the conformational dynamics of a U:U mismatch embedded within a model A-form RNA helix, flanked by all possible canonical base pair combinations. We found that the neighboring base pairs strongly influence the preferred conformational states of the U:U mismatch. However, the mismatch still regularly samples the less favored conformations on a timescale of hundreds of nanoseconds. Contrary to previous assumptions, water-mediated conformations are not universally the most stable conformational states for isolated U:U mismatches as some of the variants distinctly prefer the direct H-bonding while others destabilize the U:U mismatch altogether. Our results strongly suggest that the presence of a U:U mismatch introduces local strain into the RNA helix, which can be relieved through dynamic destabilization of either the mismatch itself or the flanking canonical base pairs, occasionally forming a shifting \"bubble\" of instability. These findings advance our understanding of U:U mismatch behavior in RNA and reveal a complex interplay between local sequence context, structural stability, and RNA dynamics.