Label-Free Live-Cell Imaging improves Mode of Action Classification
Label-Free Live-Cell Imaging improves Mode of Action Classification
Forsgren, E.; Rietdijk, J.; Holmberg, D.; Johansson, M. M.; Carreras-Puigvert, J.; Trygg, J.; Lovell, G.; Spjuth, O.; Jonsson, P.
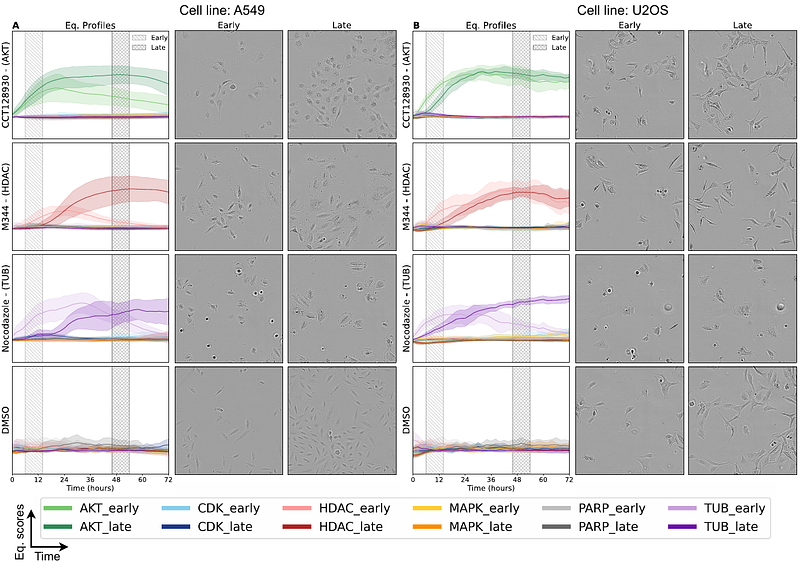
AbstractMorphological profiling is a powerful method for identifying the modes of action (MOAs) of chemical compounds. However, most approaches rely on fixed-cell assays that capture only a single time point, missing the dynamic nature of cellular responses. While live-cell imaging captures these temporal effects, its potential for MOA classification using high-dimensional features remains unexplored. We strategically convert large-scale time-series image data (>82,000 images) into interpretable phenotypic trajectories and show that label-free live-cell imaging captures meaningful temporal phenotypic signatures that improve MOA classification compared to single-time-point analyses. The results are consistent across two cell lines and six MOA groups. This workflow enables mechanistic insight from live-cell images and supports the growing interest in label-free methods, which offer advantages in preparation time and cost, and preserve the native state of cells. Our findings demonstrate that live-cell imaging, combined with data-driven analysis, offers a powerful tool for high-throughput drug screening.