Design of solubly expressed miniaturized SMART MHCs
Design of solubly expressed miniaturized SMART MHCs
White, W. L.; Bai, H.; Kim, C. J.; Jude, K. M.; Sun, R.; Guerrero, L.; Han, X.; Chen, X. T.; Chaudhuri, A.; Bonzanini, J. E.; Sun, Y.; Onwuka, A. E.; Wang, N.; Wang, C.; Li, X.; Goreshnik, I.; Allen, A.; Levine, P. M.; Kueh, H. Y.; Jewett, M. C.; Sgourakis, N.; Achour, A.; Garcia, K. C.; Baker, D.
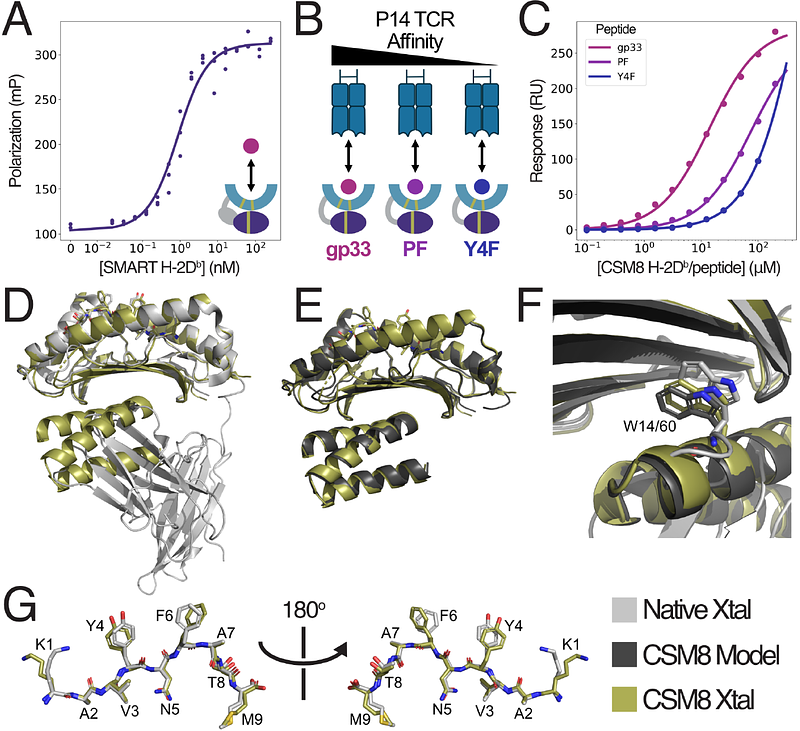
AbstractThe precise recognition of specific peptide-MHC (pMHC) complexes by T-cell receptors (TCRs) plays a key role in infectious disease, cancer and autoimmunity. A critical step in many immunobiological studies is the identification of T-cells expressing TCRs specific to a given pMHC antigen. However, the intrinsic instability of empty class-I MHCs limits their soluble expression in Escherichia coli (E. coli) and makes it very difficult to characterize even a small fraction of possible pMHC/TCR interactions. To overcome this limitation, we designed small proteins which buttress the peptide binding groove of class I MHCs, replacing {beta}2-microglobulin ({beta}2m) and the heavy chain 3 domain, and enable soluble expression of both H-2Db and A*02:01 in E. coli. We demonstrate that these soluble, monomeric, antigen-receptive, truncated (SMART) MHCs retain both peptide- and TCR-binding specificity, and that peptide-bound structures of both allomorphs are similar to their full-length, native counterparts. With extension to the majority of HLA alleles, SMART MHCs should be broadly useful for probing the T-cell repertoire in approaches ranging from yeast display to T-cell staining.