An expanded reference catalog of translated open reading frames for biomedical research
An expanded reference catalog of translated open reading frames for biomedical research
Chothani, S.; Ruiz-Orera, J.; Tierney, J. A. S.; Clauwaert, J.; Deutsch, E. W.; Alba, M. M.; Aspden, J. L.; Baranov, P. V.; Bazzini, A. A.; Bruford, E. A.; Brunet, M. A.; Cardon, T.; Carvunis, A.-R.; Casola, C.; Choudhary, J. S.; Dean, K.; Faridi, P.; Fierro-Monti, I.; Fournier, I.; Frankish, A.; Gerstein, M.; Hubner, N.; Jiang, Y.; Kellis, M.; Kok, L. W.; Martinez, T. F.; Menschaert, G.; Ni, P.; Orchard, S.; Roucou, X.; Rozowsky, J.; Salzet, M.; Siragusa, M.; Slavoff, S.; Swirski, M. I.; Valen, E.; Vizcaino, J. A.; Wacholder, A.; Wu, W.; Xie, Z.; Yang, Y. T.; Moritz, R. L.; Mudge, J.; van Hee
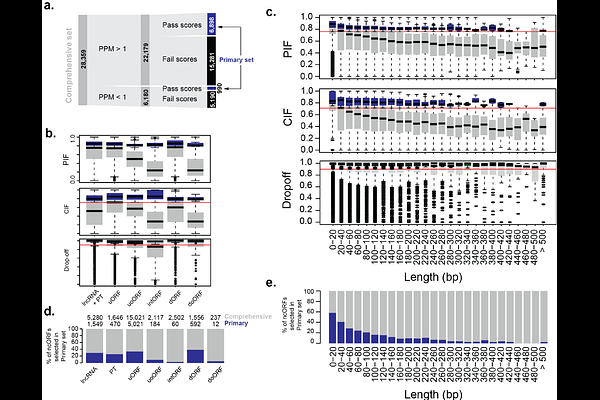
AbstractNon-canonical (i.e., unannotated) open reading frames (ncORFs) have until recently been omitted from reference genome annotations, despite evidence of their translation, limiting their incorporation into biomedical research. To address this, in 2022, we initiated the TransCODE consortium and built the first community-driven consensus catalog of human ncORFs, which was openly distributed to the research community via Ensembl-GENCODE. While this catalog represented a starting point for reference ncORF annotation, major technical and scientific issues remained. In particular, this initial catalogue had no standardized framework to judge the evidence of translation for individual ncORFs. Here, we present an expanded and refined catalog of the human reference annotation of ncORFs. By incorporating more datasets and by lifting constraints on ORF length and start-codon, we define a comprehensive set of 28,359 ncORFs that is nearly four times the size of the previous catalog. Furthermore, to aid users who wish to work with ncORFs with the strongest and most reproducible signals of translation, we utilized a data-driven framework (i.e. translation signature scores) to assess the accumulated evidence for any individual ncORF. Using this approach, we derive a subset of 7,888 ncORFs with translation evidence on par with canonical protein-coding genes, which we refer to as the Primary set. This set can serve as a reliable reference for downstream analyses and validation, with a particular emphasis on high quality. Overall, this update reflects continual community-driven efforts to make ncORFs accessible and actionable to the broader research public and further iterations of the catalog will continue to expand and refine this resource.