Improving Differential Expression and Survival Analyses with Sample Specific Compartment Deconvolution.
Improving Differential Expression and Survival Analyses with Sample Specific Compartment Deconvolution.
Yurovsky, A.; Moffitt, R. A.
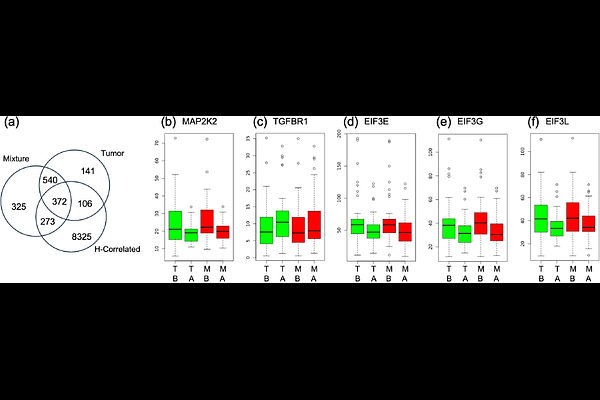
AbstractMotivation: Studies on bulk RNA-seq of tumor biopsies can yield incorrect results because varying proportions of non-tumor tissues in the samples obscure the true signal and impact the accuracy of survival and differential expression analyses. Single-cell sequencing avoids these problems, but is still too expensive in clinical settings. Other deconvolution algorithms extract tissue-specific gene expression profiles from bulk sequencing, but cannot do this on a per-sample basis. Results: We introduce SSCD - sample specific compartment deconvolution. SSCD extends non-negative matrix factorization with per-sample, per-gene constraint optimization. On simulated data, SSCD shows improvements in accuracy over existing methods. Using several real cancer datasets, we show that SSCD refines Differential Expression and survival analyses.