An organotypic in vitro model of human papillomavirus-associated precancerous lesions allowing automated cell quantification for preclinical drug testing
An organotypic in vitro model of human papillomavirus-associated precancerous lesions allowing automated cell quantification for preclinical drug testing
Köhler, R. M.; Stark, H.-J.; Martin, I.; Altmann, J.; Kalteis, M. S.; von Knebel Doeberitz, M.; Prigge, E.-S.
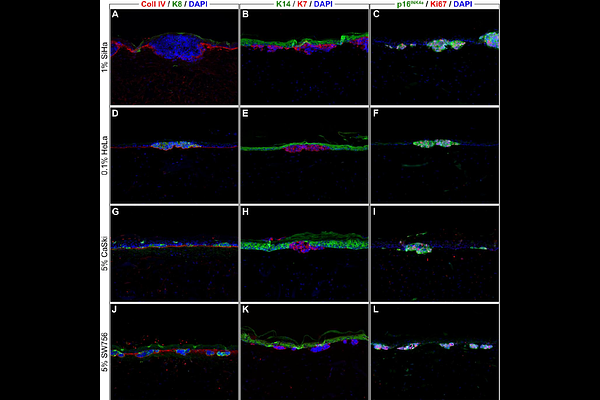
AbstractOncogenic human papillomaviruses (HPV) are causally responsible for invasive cancers and precancerous lesions. These lesions represent a considerable disease burden worldwide, yet no causally effective treatments are available. The development of HPV tumor models realistically reflecting the in vivo treatment situation is necessary for finding new effective and tissue-sparing treatments. This study aimed to establish an in vitro model of HPV-induced precancerous lesions for preclinical drug testing and to provide an automated method for quantifying cell counts to assess treatment effects in this model. To establish organotypic epithelial raft cultures (OTCs) as a model of HPV-induced precancerous lesions, we cultivated HPV-transformed cervical cancer cell lines SiHa, CaSki, HeLa, and SW756 in conjunction with primary human keratinocytes on a dermal equivalent and evaluated the impact of different cultivation variables. To demonstrate suitability of our model for preclinical drug application studies, we applied 5-fluorouracil, sinecatechins, and cisplatin either onto the air-exposed surface or to the growth medium, mimicking topical and systemic drug administration routes. We then developed a machine learning-based approach to quantify cell counts reflecting treatment effects in the established OTCs. We successfully optimized a durable in vitro model of HPV-induced precancerous lesions. The model enabled monitoring of treatment effects in a three-dimensional context with differentiated consideration of the targeted HPV-transformed cells and surrounding normal epithelium. We demonstrated that extreme gradient boosted tree classifiers (XGBoost) can be successfully used to distinguish the number of tumor cells, normal keratinocytes, and degraded cells with high accuracy (81.8%; P=2*10^-6). Critically, quantification results plausibly reflected microscopical observations and gave a fine-grained picture of time- and dose-dependent treatment effects. The established model in combination with automated cell quantification can serve as a valuable tool in analyzing major preclinical endpoints in in vitro studies, such as target cell treatment efficacy, potential side effects, and treatment schedule optimization.