Three novel genomes broaden the wild side of the Capsicum pangenome
Three novel genomes broaden the wild side of the Capsicum pangenome
Papastolopoulou, C.; Nieuwenhuis, R.; Warris, S.; Bakker, L. V.; van Haarst, J.; Cordewener, J.; Hesselink, T.; van den Broeck, H. C.; van Dooijeweert, W.; de Jong, H.; Chunwongse, J.; Diaz Trivino, S.; Schijlen, E.; de Ridder, D.; Smit, S.; Peters, S.
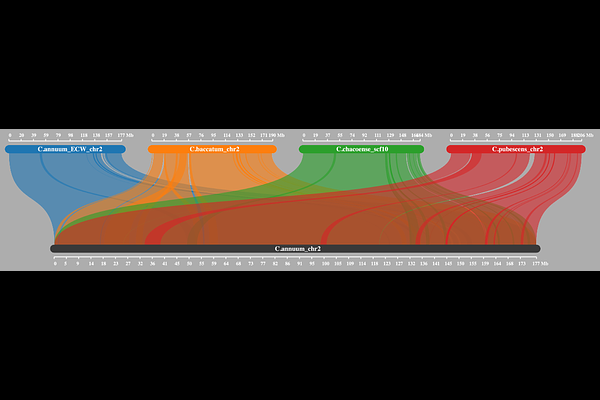
AbstractThis study presents three genome assemblies within the Capsicum genus, enabling comprehensive comparative analyses for the Annuum and Baccatum complexes within the genus. We produced highly continuous assemblies of the nuclear genomes and complete chloroplast assemblies. Subsequent genome annotation identified 34,580 genes in non-pungent C. annuum cv. ECW, and 32,704 and 33,994 genes in pungent C. chacoense and C. galapagoense, respectively. These assemblies, including the first complete genomes for C. chacoense and C. galapagoense, provide additional genomic resolution within the Capsicum genus. The novel genomes were analyzed within a pangenomic framework, integrating 16 Capsicum genomes across the Annuum, Baccatum, and Pubescens complexes. Homology grouping was used to identify core, accessory and unique genes and showed a wide spectrum of genetic diversity, particularly in homology groups exclusive to C. chacoense and C. galapagoense. Out of 79,267 homology groups identified, 13% were core groups, present in all accessions, corresponding to approximately 30% of core genes per genome. Comparative analyses revealed distinct species and genus specific genomic characteristics. Additionally, we used the graph pangenome to illustrate locus-level exploration by examining the Pun1 locus associated with capsaicinoid biosynthesis, identifying multiple Pun1-like genes including their genomic position and homology information. The integration of these new resources into a dynamic Capsicum pangenome framework provides a versatile platform for extracting genetic information relevant to both fundamental research and breeding applications.