Phase-Specific Antibiotic Resistance Mechanisms in an Escherichia coli B Strain
Phase-Specific Antibiotic Resistance Mechanisms in an Escherichia coli B Strain
Terrazas-Lopez, M.; Aitken, V.; Zeczycki, T. N.; Koculi, E.
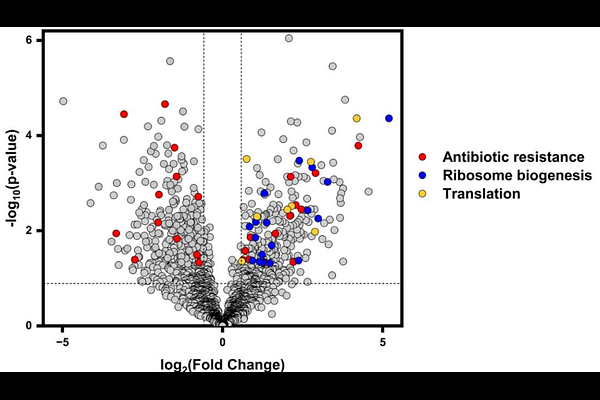
AbstractThe majority of antibiotics developed to date target the fast-growing phase of bacteria, which typically occurs during active infection and resembles the exponential growth phase of laboratory-grown cultures. However, many pathogenic bacteria in the human body occupy environments where nutrients are limited and persist in a low-metabolic state, mirroring the stationary phase observed in laboratory-grown cultures. Bacteria in this latter phase are generally more resistant to antibiotics and are often responsible for recurrent infections. A comprehensive understanding of metabolic processes in both the exponential and stationary phases of bacterial growth could facilitate the development of phase-specific antibiotics. In this study, we adopted an untargeted proteomics approach to establish similarities and differences in the exponential and stationary phase proteome landscapes in BLR (DE3) cells, an Escherichia coli B strain. Our findings are consistent with previous studies, which show that metabolic processes such as translation and ribosome biogenesis decrease during the stationary phase compared to the fast-growing exponential phase, whereas tricarboxylic acid cycle activity increases. Importantly, our data reveal that distinct antibiotic resistance pathways are activated during the exponential and stationary phases, underscoring the potential for developing phase-specific antibiotic therapies.