Biophysical characterization of Eag chaperones suggests the mechanism of effector transmembrane domain release
Biophysical characterization of Eag chaperones suggests the mechanism of effector transmembrane domain release
Van Schepdael, M.; Asakereh, I.; Colautti, J.; Gierys, A. J.; Ahmad, S.; Khajehpour, M.; Whitney, J. C.; Prehna, G.
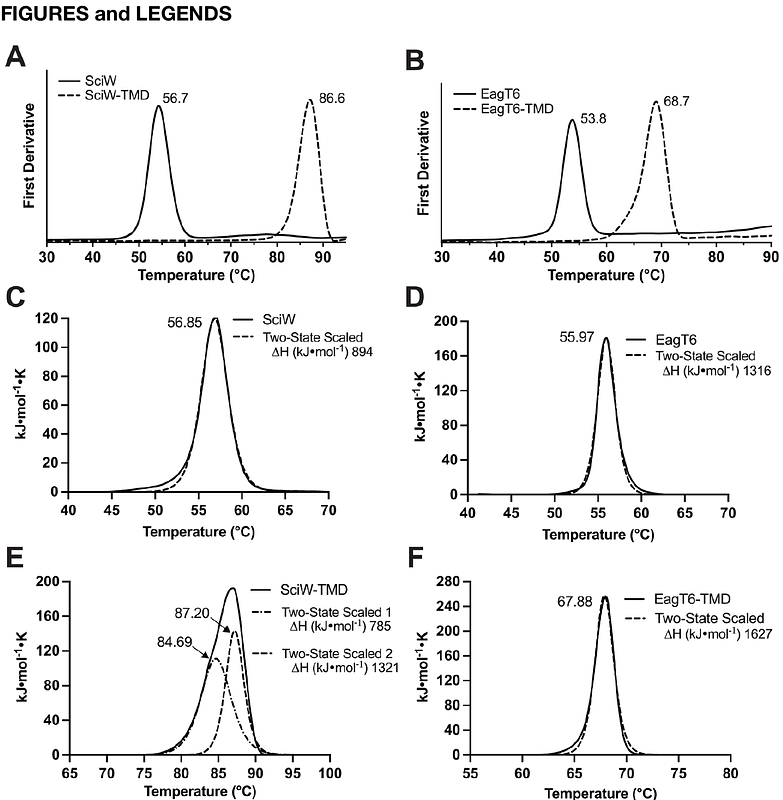
AbstractThe type VI secretion system (T6SS) is a dynamic protein nanomachine found in Gram-negative bacteria that secretes toxic effectors into prey-cells. For secretion, effectors require chaperones or adaptors for proper loading onto the T6SS. Effector associated genes (Eags) are a family of T6SS chaperones that stabilize N-terminal transmembrane domains (TMDs) found in thousands of effectors. Eags are essential for secretion and inhibit effector TMDs from prematurely adopting a membrane-penetrative conformation. However, the mechanism of TMD release from its cognate Eag chaperone is unknown. Here, we take a biochemical and biophysical approach to probe the mechanism of TMD binding and dissociation from Eag chaperones. Using steady-state fluorescence, stopped-flow measurements, and bacterial competition assays, we compare the thermodynamics, kinetics, and in vivo chaperone function of wild-type and point-variant Eag-TMD complexes. Additionally, we solve an X-ray crystal structure of an Eag-TMD point-variant complex that captures an intermediate state of TMD release. Our data reveals the molecular features and specific residue contacts necessary for TMD binding and demonstrates the Eag conformational change required to initiate rapid release of the TMD. Overall, our work details the stability of Eag-TMD complexes and the energetic pathway for the dissociation of effector TMDs from their Eag chaperones.