Towards the Development of Isoenergetic Peptide Nucleic Acid Based Probes Targeting Double-Stranded RNAs Through Enhancing Sequence-Specific Stacking Interactions
Towards the Development of Isoenergetic Peptide Nucleic Acid Based Probes Targeting Double-Stranded RNAs Through Enhancing Sequence-Specific Stacking Interactions
Lu, R.; Xi, K.; Lin, R.; Krishna, M. S.; Chen, Y.; Zhan, X.; zeng, h.; Li, X.; Fan, S.; Zhou, H.; Wu, W.; Lian, Y.; Dai, Y.; Zhu, L.; Li, G.; Chen, G.
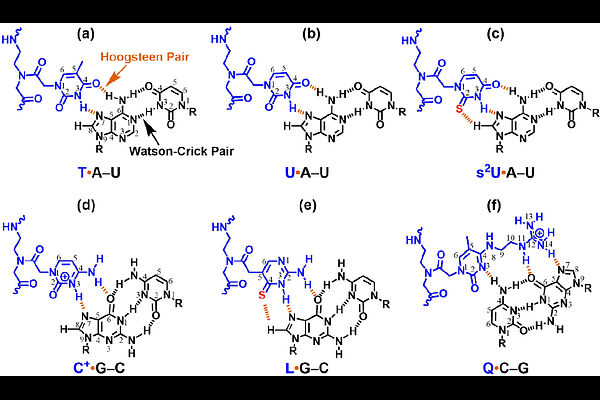
AbstractPeptide nucleic acid (PNA), a synthetic nucleic acid analog, exhibits substantial potential in biotechnology and therapeutic applications due to its high binding affinity and nuclease/protease resistance. Chemically modified PNAs capable of forming stable triplex structures with double-stranded RNAs (dsRNAs) under near-physiological conditions further expand their utility by enabling sequence-specific precise targeting and probing of functional RNA structural motifs. However, the presence of inverted Watson-Crick pairs (C-G and U-A) may significantly weaken the triplex formation of the dsRNA-binding PNAs (dbPNAs). Our previous work demonstrated that dbPNA P3 (composed of L, T, and Q monomers for the recognition of G-C, A-U, and C-G base pairs, respectively) can stimulate ribosomal frameshifting by binding to rHP2, a model RNA hairpin structure in an mRNA, albeit with suboptimal efficiency, due to its significantly weakened Q C-G triple formation. We hypothesize that incorporating s2U adjacent to Q may offer unique stacking and hydrogen bonding interactions facilitating the development of isoenergetic dbPNA-based probes binding toward dsRNAs with varied sequences. In this study, we investigate how incorporating s2U adjacent to Q residues in P3 influences its binding to rHP2. Bio-layer interferometry (BLI) and non-denaturing polyacrylamide gel electrophoresis (PAGE) analyses demonstrate that substituting T with s2U at the N-terminal position adjacent to Q (P3-2QT) enhances binding affinity by ~10-fold compared to unmodified P3, whereas C-terminal substitution (P3-TQ2) yields only a 2-fold improvement. Consistent with these findings, a cell-free dual-luciferase reporter assay reveals that P3-2QT significantly increases ribosomal frameshifting efficiency compared to P3 and P3-TQ2. Molecular dynamics simulations further indicate that P3-2QT maintains enhanced PNA-PNA stacking stability, particularly between s2U3 and Q4, suggesting a structural basis for its superior activity. Intriguingly, analogous s2U substitution in the P5 oligomer with the Q replaced by L does not confer a comparable enhancement in binding to the target RNA (rHP1) and frameshifting stimulation, highlighting the context-dependent nature of this modification. To assess the broader applicability of the s2U-Q motif, we examined its effect in dbPNAs targeting the RNA panhandle structure of influenza virus A and precursor microRNA-21, respectively. Both PAGE and BLI data confirm that s2U incorporation improves binding affinity, reinforcing the generality of this strategy. These findings underscore the potential of sequence-dependent uracil thiolation in optimizing triplex-forming dsRNA-binding PNAs, warranting further exploration of modified nucleobase designs to enhance their binding and functional properties.