AI assisted design of ligands for Lipocalin-2
AI assisted design of ligands for Lipocalin-2
Sgrignani, J.; Buscarini, S.; Locatelli, P.; Guerra, C.; Furlan, A.; Chen, Y.; Zoppi, G.; Cavalli, A.
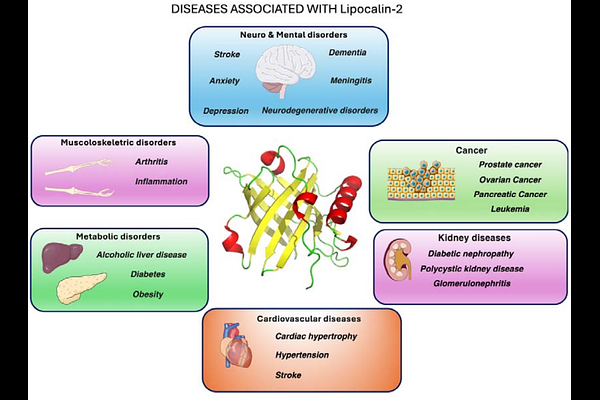
AbstractLipocalin-2 (LCN2) is an acute-phase glycoprotein whose up-regulation correlates with blood brain-barrier breakdown and neuro-inflammatory damage, making it an attractive diagnostic and therapeutic target. Here we present an end to end, AI guided workflow that rapidly generates de-novo miniproteins able to bind LCN2. Backbone scaffolds were generated with RFdiffusion, sequences were optimized with ProteinMPNN, and candidates filtered in silico using a consensus score based on AlphaFold2 confidence scores (mean interface pAE < 10) and interaction free energy calculated by Prodigy before experimental screening. From 10,000 designed sequences, five were expressed and purified from E. coli. Biolayer interferometry (BLI) identified a lead construct, MinP 2, that bound to LCN2 with dissociation constant of Kd = 0.7 nM. Furthermore, structural modeling suggests that binding is primarily mediated by hydrogen bonds between backbone elements. These findings demonstrate that a fully computational generative pipeline can deliver nanomolar LCN2 binders in a single \'design build test\' cycle. MinP 2 offers a promising starting point for affinity maturation, structural elucidation, and in-vivo evaluation as an imaging probe or antagonist of LCN2-mediated signaling.