Intra-Strain Genetic Heterogeneity in Toxoplasma gondii ME49: Oxford Nanopore Long-Read Sequencing Reveals Copy Number Variation in the ROP8-ROP2A Locus
Intra-Strain Genetic Heterogeneity in Toxoplasma gondii ME49: Oxford Nanopore Long-Read Sequencing Reveals Copy Number Variation in the ROP8-ROP2A Locus
Gohar, Y.; Neumann, M.; Huelse, L.; Wind, D.; Mock, J.; Buchholz, K.; Helle, M.; Sorg, U. R.; Degrandi, D.; Pfeffer, K.; Dilthey, A. T.
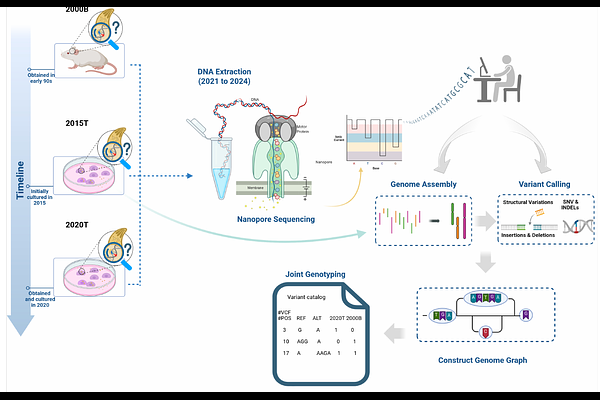
AbstractBackground: Toxoplasma gondii is an important pathogen and model organism for studying mechanisms of immune evasion and defense. Within the same strain, model organisms are typically assumed to be isogenic; for T. gondii, within-strain genetic divergence has been detected based on phenotypic changes and older molecular techniques but not characterized at the genomic level. We therefore used Oxford Nanopore long-read sequencing to characterize three independently maintained T. gondii ME49 isolates: 2015T and 2020T (obtained from ATCC and propagated in cell culture), and 2000B (propagated in mice). Results: We de novo assembled a new T. gondii ME49 reference genome and, using state of-the-art variant calling combined with pangenomic genotyping, detected variants between the sequenced isolates. Our new reference genome exceeded existing reference genomes in continuity (NG50 = 6.68 Mb versus 1.2 Mb in RefSeq) and structural accuracy, resolving all chromosomes except for a single break in the ribosomal DNA region. For isolates 2000B and 2020T, we identified 106 and 128 variants, respectively, across a final call set of 79 SNVs, 93 INDELs, and five structural variants; 24 small non-synonymous variants included genes associated with T. gondii life cycle (AP2X-8) and virulence in vivo (6-phosphogluconate dehydrogenase). A 13 kb expansion in the ROP8-ROP2A virulence locus increased the copy number of ROP2A-ROP8 genes in isolates 2000B and 2020T from three to six. Conclusions: We provide an improved T. gondii ME49 reference genome and demonstrate the potentially confounding effect of intra-strain genetic heterogeneity, highlighting the need for continuous genomic monitoring for long-term genetic identity. Keywords: Toxoplasma gondii, within-strain variation, Oxford Nanopore long-read sequencing, structural variation, genome assembly.