Multi-season analysis reveals hundreds of drought-responsive genes in sorghum
Multi-season analysis reveals hundreds of drought-responsive genes in sorghum
Cole, B.; Zhang, W.; Shi, J.; Wang, H.; Baker, C.; Varoquaux, N.; Hollingsworth, J.; Hutmacher, R.; Dahlberg, J.; Pierroz, G.; Barry, K. W.; Singan, V.; Yoshinaga, Y.; Daum, C.; Zane, M.; Blow, M.; OMalley, R.; Shu, S.; Jenkins, J. W.; Lovell, J. T.; Schmutz, J.; Taylor, J. W.; Coleman-Derr, D.; Visel, A.; Lemaux, P. G.; Purdom, E.; Vogel, J. P.
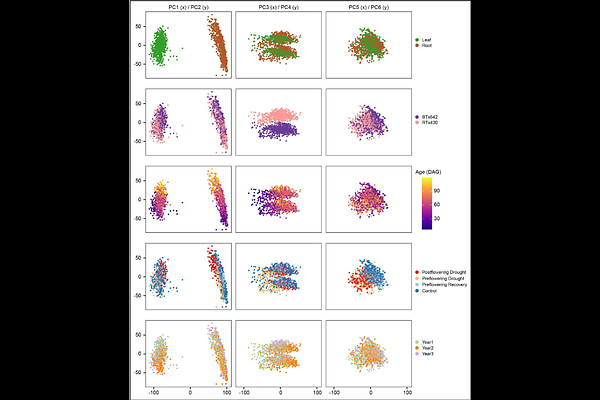
AbstractPersistent drought affects global crop production and is becoming more severe in many parts of the world in recent decades. Deciphering how plants respond to drought will facilitate the development of flexible mitigation strategies. Sorghum bicolor L. Moench (sorghum), a major cereal crop and an emerging bioenergy crop, exhibits remarkable resilience to drought. To better understand the molecular traits that underlie sorghum\'s remarkable drought tolerance, we undertook a large-scale sorghum gene expression profiling effort, totaling nearly 1,500 transcriptome profiles, across a 3-year field study with replicated plots in California\'s Central Valley. This study included time-resolved gene expression data from roots and leaves of two sorghum genotypes, BTx642 and RTx430, with different pre-flowering and post-flowering drought-tolerance adaptations under control and drought conditions. Quantification of genotype-specific drought tolerance effects was enabled by de novo sequencing, assembly, and annotation of both BTx642 and RTx430 genomes. These reference-quality genomes were used to construct a pan-gene set for characterizing conserved and genotype-specific expression. By integrating time-resolved transcriptomic responses to drought in the field across three consecutive years, we identified a set of drought-responsive genes that responded similarly in all three years of our field study. This expansive dataset represents a unique resource for sorghum and drought research communities and provides a methodological framework for the integration of multi-faceted time-resolved transcriptomic datasets.