CapTrap-seq: Advancing zebrafish transcriptomic research through high-fidelity full-length RNA sequencing
CapTrap-seq: Advancing zebrafish transcriptomic research through high-fidelity full-length RNA sequencing
Kwiatkowska, M.; Blangiewicz, M.; Madry, T.; Carbonell-Sala, S.; Guigo, R.; Uszczynska-Ratajczak, B.
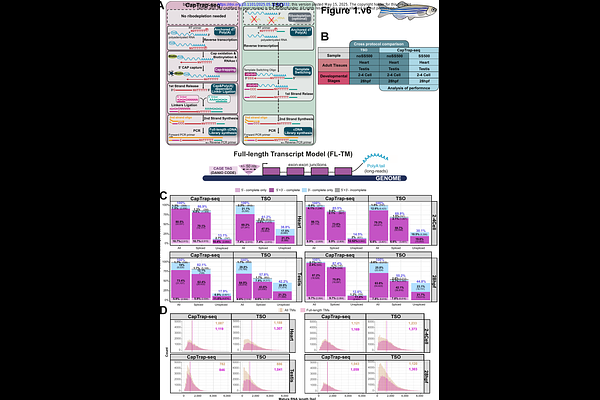
AbstractZebrafish is a valuable model organism thanks to its genetic and anatomical similarities to humans, offering a more relevant system for studying human biology and disease than in vitro or non-vertebrate models. However, its use in large-scale transcriptomic research is still limited. Most zebrafish studies focus on global expression profiling in specific developmental contexts, contributing little toward improving and expanding gene annotations. These gaps lead to inaccuracies in gene quantification and downstream functional analyses, ultimately reducing the effectiveness of zebrafish as a model system. Further challenges include ineffective ribodepletion methods and limited resources for validate transcript boundaries and splicing patterns. To overcome these challenges, we applied CapTrap-seq, a long-read sequencing method that combines Cap-trapping with oligo(dT) priming to identify 5\'-capped, full-length transcripts from both developmental and adult tissue samples. To promote detection of longer RNA molecules, we introduced a size-selection step into the CapTrap-seq protocol, which further improved detection of transcript ends without compromising its quantitative accuracy. Our results highlight the genome-agnostic nature of CapTrap-seq, enabling generation of accurate transcript models in non-mammalian systems at high throughput. CapTrap-seq also facilitates functional annotation of zebrafish genes by uncovering novel, spliced transcript isoforms with potential biological significance.