The Kink-Turn Motif: A Powerful Test for Revealing Weaknesses in RNA Force Fields
The Kink-Turn Motif: A Powerful Test for Revealing Weaknesses in RNA Force Fields
Lemmens, T.; Mlynsky, V.; Sponer, J.; Pykal, M.; Banas, P.; Otyepka, M.; Krepl, M.
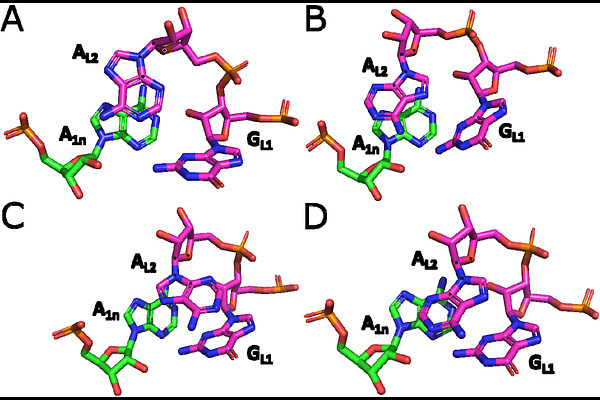
AbstractThe kink-turn is a recurrent RNA structural motif that induces a sharp bend (kink) in the A-form RNA helix. It is defined by key structural features, including consecutive sheared AG base pairs, an A-minor interaction, and multiple base-sugar interactions. Accurate representation of these densely packed non-canonical motifs in molecular dynamics simulations poses a significant challenge for contemporary force fields (FFs). Here, we present extended simulations of ribosomal kink-turn 7 (Kt-7) using a broad spectrum of pair-additive and polarizable RNA FFs. None of the tested FFs manage to flawlessly describe all the specific structural features of the Kt-7, which are described in detail in this work. Still, several FFs provide rather acceptable results and should not cause problems in simulations of larger RNAs containing a kink-turn. On aggregate, the widely used OL3 (ff99bsc0chiOL3) and polarizable AMOEBA FFs achieve the best performance. On the other hand, some more recently parametrized FF variants struggle to describe the Kt-7\'s tertiary A-minor interaction - an ubiquitous tertiary contact in RNA. This raises some concerns about the broader applicability of these FFs and suggests that they may be overfitted to small RNA model systems, such as RNA tetranucleotides. The difficulties manifest as either reversible local disruptions of the A-minor interaction and other tertiary contacts or, in some cases, irreversible unkinking of the entire motif. Based on our findings, we strongly suggest including the kink-turn motif in training and benchmarking datasets as the quintessential regression test to enhance the robustness and accuracy of RNA FF parametrization efforts.