Chronobot: Deep learning guided time-resolved cryo-EM captures molecular choreography of RecA in homology search
Chronobot: Deep learning guided time-resolved cryo-EM captures molecular choreography of RecA in homology search
Mäeots, M.-E.; Tupin, S.; Esfahani, N. M. M.; Molina, J. B. R.; Clapperton, J. A.; Amin, A.; Imbert, A.; Enchev, R. I.
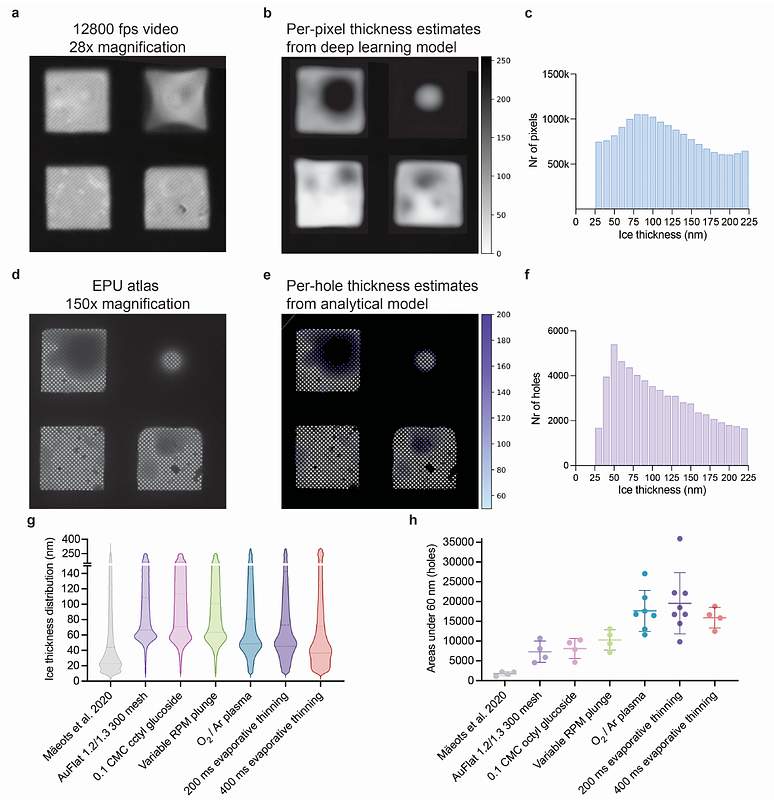
AbstractThe function of proteins and other biological macromolecules is regulated by conformational dynamics. Many functional changes take place on millisecond timescales which cannot be experimentally captured by manual sample preparation for cryo-EM. Here we introduce Chronobot, a robust, data-driven platform enabling reproducible, time-resolved cryo-EM sample preparation to visualize these transient intermediates. We quantify ice thickness, an important precondition and thus a reliable predictor of 3D reconstruction quality, using two complementary methods: a bespoke deep learning model analysing high-speed camera videos of the grid just before vitrification, and detailed ice thickness quantifications of outputs from common TEM screening workflows like the EPU software. Combining these methods enables rapid optimisation, resulting in an 11-fold improvement in cryo-EM sample quality compared to our previously reported workflow. To demonstrate the Chronobot in capturing transient reaction intermediates visualised through cryo-EM and single particle analysis we focused on RecA homology search. The RecA family of recombinases perform the essential task of rapidly scanning for homologous dsDNA sequences to initiate homologous recombination. The dynamics of these RecA-dsDNA interactions occur on millisecond timescales, limiting structural insights4. We capture time-resolved homology search intermediates at 250 milliseconds. These structures reveal the involvement of the secondary DNA binding site in initial capture of dsDNA before homology sampling occurs. We also observe three-strand homology sampling intermediates, where the homologous strand is not fully displaced, and homology is not stably bound. Our results suggest a model of how RecA-family recombinases function in early homologous recombination, by coordinating the incoming DNA between RecA\'s various DNA binding sites depending on the stage of homology search and the presence of sufficient homology. We anticipate the Chronobot method to be broadly applicable to processes which cannot be captured by manual sample preparation methods. In addition, by leveraging AI inference, our rapid user feedback mechanisms allow for per-sample optimisation of grid conditions, increasing the likelihood of success and reducing the sample requirements of each time-resolved experiment.