UV laser crosslinking uncovers novel DNA-binding pattern of CTCF
UV laser crosslinking uncovers novel DNA-binding pattern of CTCF
Stanko, C.; Oezcan, S. G.; Stengel, S.; Szymanski, L.; Steube, A.; Fischer, M.; Brioli, A.; Schenk, T.
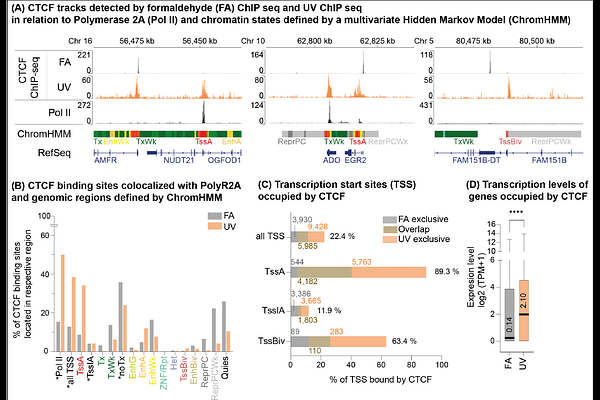
AbstractEukaryotic genomes are spatially organized to regulate gene expression, and CTCF is a key architectural protein that links chromatin topology to transcriptional control. Accurate mapping of CTCF-DNA interactions is critical for understanding gene regulation. However, conventional formaldehyde-based crosslinking in ChIP-seq biases towards long-lived protein-DNA interactions. We substituted chemical crosslinking with ultraviolet (UV) laser crosslinking (UV ChIP-seq) to capture both stable and dynamic CTCF-DNA interactions in living K-562 cells. UV ChIP-seq identified 38,706 CTCF binding sites, 70% of which were previously undetected by standard formaldehyde (FA) ChIP-seq. These UV-specific sites were enriched in active promoters, enhancers, and short-range chromatin loops, whereas formaldehyde ChIP-seq preferentially detected CTCF at topologically associated domain (TAD) boundaries and long-range loops. CTCF was detected at 85% of active transcription start sites, and when uniquely FA-identified sites were included, over 90% of active promoters were detected. De novo motif analysis also revealed noncanonical CTCF motifs at UV-specific sites, suggesting alternative binding configurations. These results expand the CTCF cistrome, redefine its role in active chromatin, and underscore the need for complementary crosslinking strategies to characterize the full spectrum of transcription factor-DNA interactions.